A guide to heat shock factors as multifunctional transcriptional regulators
- PMID: 40457168
- PMCID: PMC12366260
- DOI: 10.1111/febs.70139
A guide to heat shock factors as multifunctional transcriptional regulators
Abstract
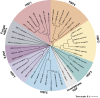
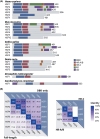
The heat shock factors (HSFs) form a family of transcription factors, which are evolutionarily conserved in eukaryotes. They are best known as transcriptional regulators of molecular chaperone genes, including those encoding heat shock proteins, in response to heat shock and other protein-damaging stresses. Since the discovery of the first HSF and its eponymous role in the heat shock response four decades ago, the currently known HSFs in vertebrates, that is, HSF1-5, HSFX, and HSFY, have been implicated in a wide array of physiological and pathological processes, including organismal development and cancer progression. To date, most studies have focused on individual HSFs, but it is becoming increasingly evident that the role of multiple HSFs and their potential crosstalk should be considered. In this review, we provide a comprehensive overview of the structures, functions, and regulation of the mammalian HSF family members and explore their interplay in biological processes. We highlight recent advancements regarding the roles of HSF family members in viral infection, cell adhesion, and spermatogenesis, and discuss the key questions to be addressed by forthcoming studies in HSF biology.
Keywords: HSE; HSF; HSP; HSR; adhesion; cancer; development; spermatogenesis; stress; transcription.
© 2025 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

