Integrative transcriptomic and proteomic analyses reveal that carbon metabolism and complement system of Madin Darby Bovine Kidney cells are affected by bovine coronavirus infection
- PMID: 40457360
- PMCID: PMC12131655
- DOI: 10.1186/s12917-025-04848-z
Integrative transcriptomic and proteomic analyses reveal that carbon metabolism and complement system of Madin Darby Bovine Kidney cells are affected by bovine coronavirus infection
Abstract
Background: Bovine coronavirus (BCoV) is a major pathogen of bovine respiratory disease, causing respiratory and enteric infections in cattle and wild ruminants. It is responsible for economic losses and threatens the health and welfare of cattle industry.
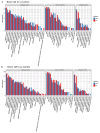
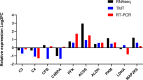
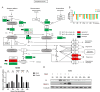
Results: In this study, a BCoV isolate, BCoV/SUWN/XHD-5, had cytopathogenic effects in Madin Darby bovine kidney (MDBK) cells and showed a high viral titer at 24 and 48 h post infection (hpi). Gene expression profiling using RNA sequencing and protein network mapping using the Tandem Mass Tag-based quantitative proteomics approach were performed in MDBK cells with BCoV infection at 24 and 48 hpi, respectively. Compared with mock-infected MDBK cells, 8,720 differentially expressed genes (DEGs) and 296 differentially expressed proteins (DEPs) were identified in BCoV infection at 24 hpi, whereas 5,838 DEGs and 747 DEPs were identified in BCoV infection at 48 hpi. Following GO annotation and KEGG enrichment analysis, most DEGs and DEPs were significantly enriched in metabolic pathways, endocytosis, ribosome and protein processing in endoplasmic reticulum, apoptosis and immune response. A correlation analysis of the proteome and transcriptome revealed that the up-regulated DEGs and DEPs were predominantly associated with metabolic pathways, apoptosis and the MAPK/TNF/Ras signaling pathway, whereas the down-regulated DEGs and DEPs were involved in complement and coagulation cascades and the Wnt signaling pathway. Importantly, BCoV decreases the mRNA and protein levels of complement component C3 in MDBK cells.
Conclusions: The findings of this study provide the first report of the integrative transcriptomic and proteomic analyses of BCoV infection in MDBK cells. It revealed a regulatory network for analyzing the mechanisms of BCoV-host interactions and provides valuable insights into the pathogenesis of BCoV.
Keywords: Bovine coronavirus; C3; Carbon metabolism; Complement systems; MDBK; Proteomics; Transcriptomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures








Similar articles
-
Integrative Transcriptomics and Proteomics Analysis Provide a Deep Insight Into Bovine Viral Diarrhea Virus-Host Interactions During BVDV Infection.Front Immunol. 2022 Mar 16;13:862828. doi: 10.3389/fimmu.2022.862828. eCollection 2022. Front Immunol. 2022. PMID: 35371109 Free PMC article.
-
Transcriptome and Proteomic Analysis Reveals Up-Regulation of Innate Immunity-Related Genes Expression in Caprine Herpesvirus 1 Infected Madin Darby Bovine Kidney Cells.Viruses. 2021 Jul 2;13(7):1293. doi: 10.3390/v13071293. Viruses. 2021. PMID: 34372499 Free PMC article.
-
Leveraging Artificial Intelligence and Gene Expression Analysis to Identify Some Potential Bovine Coronavirus (BCoV) Receptors and Host Cell Enzymes Potentially Involved in the Viral Replication and Tissue Tropism.Int J Mol Sci. 2025 Feb 4;26(3):1328. doi: 10.3390/ijms26031328. Int J Mol Sci. 2025. PMID: 39941096 Free PMC article.
-
Bovine respiratory coronavirus.Vet Clin North Am Food Anim Pract. 2010 Jul;26(2):349-64. doi: 10.1016/j.cvfa.2010.04.005. Vet Clin North Am Food Anim Pract. 2010. PMID: 20619189 Free PMC article. Review.
-
Advances in Laboratory Diagnosis of Coronavirus Infections in Cattle.Pathogens. 2024 Jun 21;13(7):524. doi: 10.3390/pathogens13070524. Pathogens. 2024. PMID: 39057751 Free PMC article. Review.
References
-
- Owens F, Di Serio F, Li S, Pallás V, Randles J, Sano. Vidalakis: virus taxonomy: ninth report of the international committee on taxonomy of viruses. Edn. 2012:1221–34.
-
- Mebus CA, White RG, Stair EL, Rhodes MB, Twiehaus MJ. Neonatal calf diarrhea: results of a field trial using a reo-like virus vaccine. Vet Med Small Anim Clin. 1972;67(2):173–4. passim. - PubMed
-
- Calderón Bernal JM, Fernández A, Arnal JL, Baselga C, Benito Zuñiga A, Fernández-Garyzábal JF, Vela Alonso AI, Cid D. Cluster analysis of bovine respiratory disease (BRD)-associated pathogens shows the existence of two epidemiological patterns in BRD outbreaks. Vet Microbiol. 2023;280:109701. - PubMed
MeSH terms
Substances
Grants and funding
- RQD2023031/Southwest Minzu University Research Startup Funds
- YBZYSC22BK07/the Science and Technology Planning Project of Yibin Vocational and Technical College
- 2024ZYD0072/Sichuan Central Guiding Local Technology Development Project
- SCCXTD-2025-18/the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System
LinkOut - more resources
Full Text Sources
Miscellaneous

