In vitro fermentation potential of diet-derived fermentable proteins of thirty-one human foods
- PMID: 40457807
- PMCID: PMC12303725
- DOI: 10.1017/S0007114525103541
In vitro fermentation potential of diet-derived fermentable proteins of thirty-one human foods
Abstract
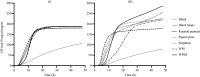
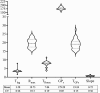
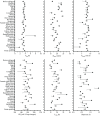
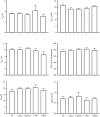
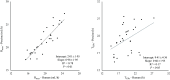
Protein fermentation in the human gut is often associated with adverse health effects. Hence, understanding the fermentation characteristics of dietary undigested proteins is important for a comprehensive nutritional value of foods. This study investigated the protein fermentation kinetics of diet-derived proteins from thirty-one different foods using an in vitro model and human faecal inoculum. The undigested diet-derived protein substrate originated from porcine ileal digesta obtained from assessment of the digestible indispensable amino acid score (DIAAS) of the foods. Significant variations in fermentation kinetic parameters, particularly in maximum gas production rate (Rmax) and time to reach cumulative gas production (GP) from the substrate (TGPs), were observed. The Rmax ranged from 15·5 (se 0·7) ml/h for wheat bran-derived to 24·5 (se 0·9) ml/h for oatmeal-derived proteins. Egg-derived proteins had the shortest TGPs (14·7 (se 0·7) h), while mushroom-derived proteins had the longest (27·6 (se 7·1) h). When foods were categorised into five groups ('animal protein', 'grains', 'legumes', 'fungi, algae and microorganisms' and 'others'), no significant differences were found in fermentation kinetics parameters. Samples were additionally incubated with porcine inoculum to assess potential donor-species effects. Human inoculum showed significantly lower Rmax, cumulative GP and microbiota turnover than porcine inoculum, indicating reduced fermentative activity. Linear regression analysis revealed correlations between human and porcine-derived inoculum only for Rmax (R2 = 0·78, P < 0·01) and TGPs (R² = 0·17, P < 0·05). These findings underscore the importance of using human inoculum in in vitro studies to better predict health implications of foods with DIAAS values.
Keywords: Gas production; Human inoculum; In vitro fermentation; Porcine model; Undigested protein.
Figures
Similar articles
-
In vitro fermentation characteristics of dietary fibers using fecal inoculum from dogs consuming commercial or grain kefir.J Anim Sci. 2025 Jan 4;103:skaf022. doi: 10.1093/jas/skaf022. J Anim Sci. 2025. PMID: 39901725
-
An in vitro model for caecal proteolytic fermentation potential of ingredients in broilers.Animal. 2023 Apr;17(4):100768. doi: 10.1016/j.animal.2023.100768. Epub 2023 Mar 9. Animal. 2023. PMID: 37011455
-
In vitro fermentation characteristics of acacia fiber using canine fecal inoculum.J Anim Sci. 2025 Jan 4;103:skaf152. doi: 10.1093/jas/skaf152. J Anim Sci. 2025. PMID: 40326307
-
Intravenous magnesium sulphate and sotalol for prevention of atrial fibrillation after coronary artery bypass surgery: a systematic review and economic evaluation.Health Technol Assess. 2008 Jun;12(28):iii-iv, ix-95. doi: 10.3310/hta12280. Health Technol Assess. 2008. PMID: 18547499
-
Nutritional interventions for survivors of childhood cancer.Cochrane Database Syst Rev. 2016 Aug 22;2016(8):CD009678. doi: 10.1002/14651858.CD009678.pub2. Cochrane Database Syst Rev. 2016. PMID: 27545902 Free PMC article.
References
-
- Russell WR, Gratz SW, Duncan SH, et al. (2011) High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr 93, 1062–1072. - PubMed
-
- Yao CK, Muir JG & Gibson PR (2016) Insights into colonic protein fermentation, its modulation and potential health implications. Aliment Pharmacol Ther 43, 181–196. - PubMed
-
- Smith EA & Macfarlane GT (1998) Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol Ecol 25, 355–368.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

