An immune-related signature based on molecular subtypes for predicting the prognosis and immunotherapy efficacy of hepatocellular carcinoma
- PMID: 40458412
- PMCID: PMC12127308
- DOI: 10.3389/fimmu.2025.1481366
An immune-related signature based on molecular subtypes for predicting the prognosis and immunotherapy efficacy of hepatocellular carcinoma
Abstract
Background: Immunotherapy has emerged as a pivotal therapeutic modality for a multitude of malignancies, notably hepatocellular carcinoma (HCC). This research endeavors to construct a prognostic signature based on immune-related genes between different HCC molecular subtypes, offer guidance for immunotherapy application, and promote its clinical practical application through immunohistochemistry.
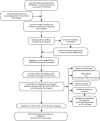
Methods: Distinguishing HCC subtypes through Gene set variation analysis and Consensus clustering analysis using the Kyoto Encyclopedia of Genes and Genome (KEGG) pathway. In the TCGA-LIHC cohort, univariate, Lasso, and multivariate Cox regression analyses were applied to construct a novel immune relevant prognostic signature. The Subtype-specific and Immune-Related Prognostic Signatures (SIR-PS) were validated in three prognostic cohorts, one immunotherapy cohort, different HCC cell lines and tissue chips. Further possible mechanism on immunotherapy was explored by miRNA-mRNA interactions and signaling pathway.
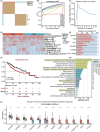
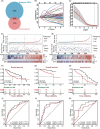
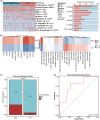
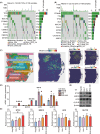
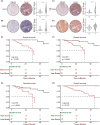
Results: This prognostic model, which was based on four critical immune-related genes, STC2, BIRC5, EPO and GLP1R, was demonstrated excellent performance in both prognosis and immune response prediction of HCC. Clinical pathological signature, tumor microenvironment and mutation analysis also proved the effective prediction of this model. Spatial transcriptome analysis shows that STC2 and BIRC5 are mainly enriched in liver cancer cells and their mRNA and protein expression levels were greater in higher malignant HCC cell lines than in the lower ones. Further validation on HCC tissue chips of this model also showed good correlation with cancer prognosis. The risk score of each patient demonstrated that the SIR-PS exhibited excellent 1 and 3-year survival prediction performance.
Conclusions: Our analysis demonstrates that the SIR-PS model serves as a robust prognostic and predictive tool for both the survival outcomes and the response to immunotherapy in hepatocellular carcinoma patients, which may shed light on promoting the individualized immunotherapy against hepatocellular carcinoma.
Keywords: biomarker; hepatocellular carcinoma; immune-related genes; immunohistochemistry; immunotherapy; prognosis.
Copyright © 2025 Sun, Jia, Liang and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

