Impact of Lactiplantibacillus plantarum on the fermentation quality, nutritional enhancement, and microbial dynamics of whole plant soybean silage
- PMID: 40458705
- PMCID: PMC12127759
- DOI: 10.3389/fmicb.2025.1565951
Impact of Lactiplantibacillus plantarum on the fermentation quality, nutritional enhancement, and microbial dynamics of whole plant soybean silage
Abstract
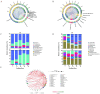
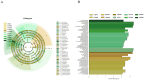
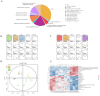
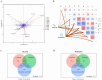
Soybean (Glycine max (L.) Merr.) is an important leguminous crop with rich nutrients and wide uses, yet soybean straw is often treated as waste in many areas without sufficient regard for its nutritional value. For the sustainable utilization of biomass resources, this study assessed the fermentation quality, microbial communities, and metabolites of whole plant soybean (WPS) silage with and without Lactiplantibacillus plantarum (LP) over different fermentation periods of 7, 15, 30, 60, and 90 days. With LP, there was a significant increase in dry matter (DM), crude protein (CP), and water-soluble carbohydrate (WSC) content of silage (p < 0.01) and a significant decrease in pH (p < 0.01). Incorporating LP into WPS silage significantly elevated lactic acid (LA) concentration, thereby improving fermentation quality. 16S rRNA gene sequencing revealed that LP inoculation significantly altered bacterial diversity and composition, notably increasing the relative abundance of Lactobacillus and promoting beneficial shifts in the microbial community during silage fermentation. Notably, LP treatment significantly promoted lysine biosynthesis, a key essential amino acid pathway, thereby contributing to the nutritional enhancement of the silage. Results showed that adding LP to WPS at ensiling can improve silage microbial community structure optimize metabolic processes, produce superior metabolites, and enhance the silage's fermentation quality and nutritional value, after 60 days of fermentation. In summary, WPS silage with LP addition could serve as a promising strategy for preserving high-protein forage silage.
Keywords: Lactiplantibacillus plantarum; bacterial community; metabolites; silage; soybean.
Copyright © 2025 Meng, Xu, Wang, Wang, Wang, Wu, Hou, Wang, Tong, Jiang and Wang.
Conflict of interest statement
YX is employed by Beidahuang Group Heilongjiang Tangyuan Farm Co., Ltd., Jiamusi, China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Ahmadi F., Lee Y. H., Lee W. H., Oh Y.-K., Park K., Kwak W. S. (2019). Long-term anaerobic conservation of fruit and vegetable discards without or with moisture adjustment after aerobic preservation with sodium metabisulfite. Waste Manag. 87, 258–267. doi: 10.1016/j.wasman.2019.02.010, PMID: - DOI - PubMed
-
- AOAC (1990). Official methods of analysis of the association of official analytical chemists. Arlington, VA: AOAC.
-
- Bai J., Ding Z., Ke W., Xu D., Wang M., Huang W., et al. (2021). Different lactic acid bacteria and their combinations regulated the fermentation process of ensiled alfalfa: ensiling characteristics, dynamics of bacterial community and their functional shifts. Microb. Biotechnol. 14, 1171–1182. doi: 10.1111/1751-7915.13785, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

