Exploring Key Genes of Glutathione Metabolism in Systemic Lupus Erythematosus Based on Mendelian Randomisation, Single-Cell RNA Sequencing and Multiple Machine Learning Approaches
- PMID: 40458850
- PMCID: PMC12130738
- DOI: 10.1049/syb2.70021
Exploring Key Genes of Glutathione Metabolism in Systemic Lupus Erythematosus Based on Mendelian Randomisation, Single-Cell RNA Sequencing and Multiple Machine Learning Approaches
Abstract
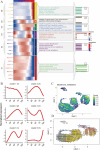
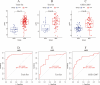
Systemic lupus erythematosus (SLE) is a complex autoimmune disease characterised by immune dysregulation leading to inflammation and organ damage. Despite the rising global incidence of SLE, its aetiology remains unclear. We applied Mendelian randomisation (MR), multi-omics integration, machine learning (ML), and SHAP to identify key metabolites and genes associated with SLE, revealing the crucial role of the glutathione pathway. MR analysis was performed on 1400 serum metabolites, revealing significant enrichment in the glutathione metabolic pathway. Single-cell RNA sequencing (scRNA-seq) data classified monocytes into Metabolism_high and Metabolism_low groups based on glutathione metabolism scores. Differentially expressed genes were analysed using GSEA, metabolic pathway activity assessment, transcription factor prediction, cellular communication analysis, and Pseudotime analysis. LASSO regression identified hub genes and machine learning models (CatBoost, XGBoost, NGBoost) were developed. The SHAP method was used to interpret these models. Expression of key genes was validated across multiple datasets. MR analysis confirmed that metabolites were enriched in the glutathione pathway, identifying nine hub genes. Machine learning models achieved AUCs of 0.85, 0.80, and 0.83 in the validation set. SHAP analysis highlighted LAP3 as the top contributing gene across all models. scRNA-seq data showed that LAP3 plays a significant role in the immune microenvironment of SLE. Validation across multiple datasets (training, validation, and GSE112087) revealed elevated LAP3 expression in PBMCs of SLE patients, with AUCs of 0.935, 0.795, and 0.817, respectively, suggesting strong diagnostic potential. Glutathione metabolism is closely associated with SLE development and LAP3 may play a key role in its progression. Both glutathione metabolism and LAP3 could serve as potential targets for SLE diagnosis and treatment.
Keywords: SHAP, multi‐omic; glutathione metabolism; machine learning; mendelian randomisation; single‐cell RNA sequencing; systemic lupus erythematosus (SLE).
© 2025 The Author(s). IET Systems Biology published by John Wiley & Sons Ltd on behalf of The Institution of Engineering and Technology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

