Functional diversification of the MADS-box gene family in fine-tuning the dimorphic transition of Talaromyces marneffei
- PMID: 40459274
- PMCID: PMC12282165
- DOI: 10.1128/msystems.00464-25
Functional diversification of the MADS-box gene family in fine-tuning the dimorphic transition of Talaromyces marneffei
Abstract
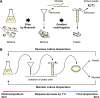
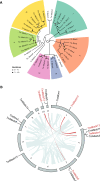
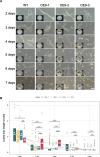
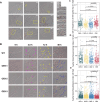
The dynamic transition between yeast and hypha is a crucial adaptive mechanism for many human pathogenic fungi, including Talaromyces marneffei, a thermodimorphic fungus responsible for causing fatal talaromycosis. In the current study, we elucidated the roles of the MADS-box family in fine-tuning the dimorphic transition in T. marneffei through functional diversification of members. Utilizing adaptive laboratory evolution, we identified an enrichment of MADS-box genes in mutants deficient in yeast-to-mycelium transition. Further phylogenetic analyses revealed a significant expansion of the MADS-box gene family within T. marneffei. Functional genetic manipulations revealed that overexpression of mads9, as opposed to its paralog mads10, effectively delayed the hypha-to-yeast transition. Through integrating RNA sequencing and chromatin immunoprecipitation sequencing, we demonstrated that mads9 and the previously characterized madsA (mads7) modulated the rate of hypha-to-yeast conversion by orchestrating metabolic pathways and membrane dynamics, respectively, with mutual regulation via shared target genes. Our findings illuminated the distinct functional roles of the MADS-box family in regulating dimorphic transitions in T. marneffei, offering new insights into fungal adaptability.
Importance: The dimorphic transition between yeast and hyphal forms in Talaromyces marneffei is a critical adaptive mechanism that underpins its pathogenicity, particularly in response to environmental cues such as temperature. In this study, we elucidated the role of the MADS-box transcription factor family and discovered that its members collaboratively regulate dimorphic transitions by assuming distinct roles in the morphogenesis, enhancing the understanding of the thermal adaptation of T. marneffei and the functional roles of the MADS-box gene family outside the plant.
Keywords: ChIP-seq; MADS-box; RNA-seq; Talaromyces marneffei; dimorphism transition; fungal adaptation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Signature gene expression reveals novel clues to the molecular mechanisms of dimorphic transition in Penicillium marneffei.PLoS Genet. 2014 Oct 16;10(10):e1004662. doi: 10.1371/journal.pgen.1004662. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25330172 Free PMC article.
-
Genome-wide identification, characterization, and expression analysis of the MADS-box gene family in grass pea (Lathyrus sativus) under salt stress conditions.BMC Genomics. 2025 May 21;26(1):519. doi: 10.1186/s12864-025-11661-3. BMC Genomics. 2025. PMID: 40399794 Free PMC article.
-
MADS-Box Transcription Factor MadsA Regulates Dimorphic Transition, Conidiation, and Germination of Talaromyces marneffei.Front Microbiol. 2018 Aug 7;9:1781. doi: 10.3389/fmicb.2018.01781. eCollection 2018. Front Microbiol. 2018. PMID: 30131782 Free PMC article.
-
Accuracy of rapid diagnosis of Talaromyces marneffei: A systematic review and meta-analysis.PLoS One. 2018 Apr 5;13(4):e0195569. doi: 10.1371/journal.pone.0195569. eCollection 2018. PLoS One. 2018. PMID: 29621346 Free PMC article.
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
References
-
- Seidel D, Wurster S, Jenks JD, Sati H, Gangneux J-P, Egger M, Alastruey-Izquierdo A, Ford NP, Chowdhary A, Sprute R, Cornely O, Thompson GR 3rd, Hoenigl M, Kontoyiannis DP. 2024. Impact of climate change and natural disasters on fungal infections. Lancet Microbe 5:e594–e605. doi: 10.1016/S2666-5247(24)00039-9 - DOI - PubMed
-
- Greenspan SE, Roznik EA, Edwards L, Duffy R, Berger L, Bower DS, Pike DA, Schwarzkopf L, Alford RA. 2023. Constant-temperature predictions underestimate growth of a fungal amphibian pathogen under individual host thermal profiles. J Therm Biol 111:103394. doi: 10.1016/j.jtherbio.2022.103394 - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
