Polygenic modifiers impact penetrance and expressivity in telomere biology disorders
- PMID: 40459934
- PMCID: PMC12352891
- DOI: 10.1172/JCI191107
Polygenic modifiers impact penetrance and expressivity in telomere biology disorders
Abstract
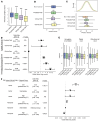
BACKGROUNDTelomere biology disorders (TBDs) exhibit incomplete penetrance and variable expressivity, even among individuals harboring the same pathogenic variant. We assessed whether common genetic variants associated with telomere length combine with large-effect variants to impact penetrance and expressivity in TBDs.METHODSWe constructed polygenic scores (PGS) for telomere length in the UK Biobank to quantify common variant burden and assessed the PGS distribution across patient cohorts and biobanks to determine whether individuals with severe TBD presentations have increased polygenic burden causing short telomeres. We also characterized rare TBD variant carriers in the UK Biobank.RESULTSIndividuals with TBDs in cohorts enriched for severe pediatric presentations have polygenic scores predictive of short telomeres. In the UK Biobank, we identified carriers of pathogenic TBD variants who were enriched for adult-onset manifestations of TBDs. Unlike individuals in disease cohorts, the PGS of adult carriers did not show a common variant burden for shorter telomeres, consistent with the absence of childhood-onset disease. Notably, TBD variant carriers were enriched for idiopathic pulmonary fibrosis diagnoses and telomere length PGS stratified pulmonary fibrosis risk. Finally, common variants affecting telomere length were enriched in enhancers regulating known TBD genes.CONCLUSIONCommon genetic variants combined with large-effect causal variants to impact clinical manifestations in rare TBDs. These findings offer a framework for understanding phenotypic variability in other presumed monogenic disorders.FUNDINGThis work was supported by NIH grants R01DK103794, R01HL146500, R01CA265726, R01CA292941, and the Howard Hughes Medical Institute.
Keywords: Bone marrow; Genetics; Hematology; Hematopoietic stem cells; Stem cells; Telomeres.
Conflict of interest statement
Figures



Comment in
- Genetic risk in telomere biology disorders: it adds up doi: 10.1172/JCI195921
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

