FGFR3-induced Y158 PARP1 phosphorylation promotes PARP inhibitor resistance via BRG1/MRE11-mediated DNA repair in breast cancer models
- PMID: 40460005
- PMCID: PMC12259256
- DOI: 10.1172/JCI173757
FGFR3-induced Y158 PARP1 phosphorylation promotes PARP inhibitor resistance via BRG1/MRE11-mediated DNA repair in breast cancer models
Abstract
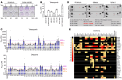
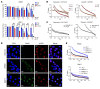
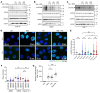
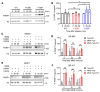
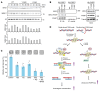
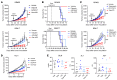
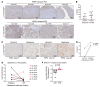
Poly(ADP-ribose) polymerase (PARP) inhibitors (PARPis) are used to treat BRCA-mutated (BRCAm) cancer patients; however, resistance has been observed. Therefore, biomarkers to indicate PARPi resistance and combination therapy to overcome that are urgently needed. We identified a high prevalence of activated FGF receptor 3 (FGFR3) in BRCAm triple-negative breast cancer (TNBC) cells with intrinsic and acquired PARPi resistance. FGFR3 phosphorylated PARP1 at tyrosine 158 (Y158) to recruit BRG1 and prolong chromatin-loaded MRE11, thus promoting homologous recombination (HR) to enhance PARPi resistance. FGFR inhibition prolonged PARP trapping and synergized with PARPi in vitro and in vivo. High-level PARP1 Y158 phosphorylation (p-Y158) positively correlated with PARPi resistance in TNBC patient-derived xenograft models, and in PARPi-resistant TNBC patient tumors. These findings reveal that PARP1 p-Y158 facilitates BRG1-mediated HR to resolve the PARP-DNA complex, and PARP1 p-Y158 may indicate PARPi resistance that can be relieved by combining FGFR inhibitors (FGFRis) with PARPis. In summary, we show that FGFRi restores PARP trapping and PARPi antitumor efficacy in PARPi-resistant breast cancer by decreasing HR through the PARP1 p-Y158/BRG1/MER11 axis, suggesting that PARP1 p-Y158 is a biomarker for PARPi resistance that can be overcome by combining FGFRis with PARPis.
Keywords: Breast cancer; Drug therapy; Oncology; Protein kinases; Therapeutics.
Conflict of interest statement
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

