Context-Aware Biosensor Design Through Biology-Guided Machine Learning and Dynamical Modeling
- PMID: 40460061
- PMCID: PMC12186671
- DOI: 10.1021/acssynbio.4c00894
Context-Aware Biosensor Design Through Biology-Guided Machine Learning and Dynamical Modeling
Abstract
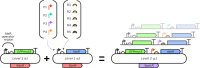
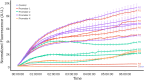
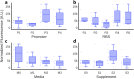
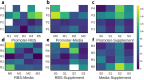
Addressing the challenge of achieving a global circular bioeconomy requires efficient and robust bio-based processes operating at different scales. These processes should also be competitive replacements for the production of chemicals currently obtained from fossil resources, as well as for the production of new-to-nature compounds. To that end, genetic circuits can be used to control cellular behavior and are instrumental in developing efficient cell factories. Whole-cell biosensors harbor circuits that can be based on allosteric transcription factors (TFs) to detect and elicit a response depending on the target molecule concentrations. By modifying regulatory elements and testing various genetic components, the responsive behavior of genetic biosensors can be finely tuned and engineered. While previous models have described and characterized the behavior of naringenin biosensors, additional data and resources are required to predict their dynamic response and performance in different contexts, such as under various gene expression regulatory elements, media, carbon sources, or media supplements. Tuning these conditions is pivotal in optimizing biosensor design for applications operating in varying conditions, such as fermentation processes. In this study, we assembled a library of FdeR biosensors, characterized their performance under different conditions, and developed a mechanistic model to describe their dynamic behavior under reference conditions, which guided a machine learning-based predictive model that accounts for context-dependent dynamic parameters. Such a Design-Build-Test-Learn (DBTL) pipeline allowed us to determine optimal condition combinations for the desired biosensor specifications, both for automated screening and dynamic regulation. The findings of this work contribute to a deeper understanding of whole-cell biosensors and their potential for precise measurement, screening, and dynamic regulation of engineered production pathways for valuable molecules.
Keywords: biosensor; context dependence; dynamical modeling; genetic circuit; scientific machine learning; synthetic biology.
Figures

Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

