Microbial diversity of coastal microbial mats formations in karstic habitats from the Yucatan Peninsula, Mexico
- PMID: 40460360
- PMCID: PMC12133189
- DOI: 10.1371/journal.pone.0325200
Microbial diversity of coastal microbial mats formations in karstic habitats from the Yucatan Peninsula, Mexico
Abstract
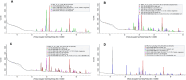
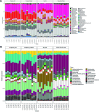
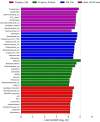
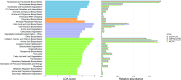
In this study, we report for the first time an exploration of the physicochemical characteristics and the prokaryotic diversity of three different types of microbial mats from karstic habitats located in Sisal, Progreso and Ría Lagartos, in the Yucatan Peninsula, Mexico. Our results showed that lift-off mats were found in the lower salinity (2.2%) area (Sisal), while flat and pustular mats were detected in hypersaline (6-9%) sites (Progreso and Ría Lagartos). Notably, some of these microbial mat structures were in close proximity to mangrove forest ecosystems with both degraded and restored regimes. XRD analysis revealed different mineral compositions of the mats; however, aragonite, calcite, and halite were commonly found in all samples studied. High-throughput sequencing of the 16S rRNA gene identified differences in microbial communities across the different mat types, and statistical analyses revealed that salinity, redox potential, and temperature were significant factors in explaining the variance of the prokaryotic assemblages. Microbial groups identified in this study include those known to be important in the biogeochemical cycling of key elements, such as carbon, nitrogen, and sulfur. Interestingly, the community composition of flat and pustular mats from Progreso was similar, with Bacteroidia, Anaerolineae, and Phycisphaerae being the most abundant microbial groups in flat mats; and Bacteroidia, Anaerolineae, and Alphaproteobacteria dominating pustular mats. By contrast, flat mats from Ría Lagartos were dominated by Halobacteria, Cyanobacteria and Bacteroidota, while Bacteroidia, Gammaproteobacteria and Cyanobacteria dominated lift-off mats from Sisal. This work contributes to understanding the distribution, physicochemical characteristics and microbial diversity of coastal microbial mats, providing valuable new insights into microbial mats that develop in karstic ecosystems. This information is relevant to ongoing and future efforts to manage and preserve coastal ecosystems in the Yucatan Peninsula.
Copyright: © 2025 Cadena et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Bacterial and Archaeal Diversity in Hypersaline Cyanobacterial Mats Along a Transect in the Intertidal Flats of the Sultanate of Oman.Microb Ecol. 2018 Feb;75(2):331-347. doi: 10.1007/s00248-017-1040-9. Epub 2017 Jul 24. Microb Ecol. 2018. PMID: 28736793
-
Analysis of bacterial and archaeal diversity in coastal microbial mats using massive parallel 16S rRNA gene tag sequencing.ISME J. 2011 Nov;5(11):1701-12. doi: 10.1038/ismej.2011.52. Epub 2011 May 5. ISME J. 2011. PMID: 21544102 Free PMC article.
-
Do restoration strategies in mangroves recover microbial diversity? A case study in the Yucatan peninsula.PLoS One. 2024 Aug 16;19(8):e0307929. doi: 10.1371/journal.pone.0307929. eCollection 2024. PLoS One. 2024. PMID: 39150908 Free PMC article.
-
Prokaryotic diversity and biogeochemical characteristics of benthic microbial ecosystems at La Brava, a hypersaline lake at Salar de Atacama, Chile.PLoS One. 2017 Nov 15;12(11):e0186867. doi: 10.1371/journal.pone.0186867. eCollection 2017. PLoS One. 2017. PMID: 29140980 Free PMC article.
-
Molecular ecology of microbial mats.FEMS Microbiol Ecol. 2014 Nov;90(2):335-50. doi: 10.1111/1574-6941.12408. Epub 2014 Aug 28. FEMS Microbiol Ecol. 2014. PMID: 25109247 Review.
References
-
- Stal LJ. Coastal microbial mats: the physiology of a small-scale ecosystem. S Afr J Bot. 2001;67(3):399–410. doi: 10.1016/s0254-6299(15)31156-x - DOI
-
- Stal LJ. Microbial mats in coastal environments. In: Stal LJ, Caumette P, editors. Microbial Mats. Berlin, Heidelberg: Springer Berlin Heidelberg; 1994. pp. 21–32. doi: 10.1007/978-3-642-78991-5_2 - DOI
-
- Gerdes G. What are Microbial Mats? In: Seckbach J, Oren A, editors. Microbial Mats. Dordrecht: Springer Netherlands; 2010. pp. 3–25. doi: 10.1007/978-90-481-3799-2_1 - DOI
-
- Hoffman P, Logan BW, Gebelein CD. Algal mats, cryptalgal fabrics, and structures, hamelin pool, Western Australia. AAPG Bulletin. 1972;56:628–628. doi: 10.1306/819a3f6a-16c5-11d7-8645000102c1865d - DOI
-
- Play Ford PE, Cockbain AE. Chapter 8.2 modern algal stromatolites at hamelin pool, a hypersaline barred basin in shark bay, Western Australia. Developments in Sedimentology. Elsevier. 1976. p. 389–411. doi: 10.1016/s0070-4571(08)71147-6 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

