Sodium azide mutagenesis induces a unique pattern of mutations
- PMID: 40460371
- PMCID: PMC12162104
- DOI: 10.1371/journal.pgen.1011634
Sodium azide mutagenesis induces a unique pattern of mutations
Abstract
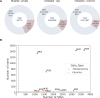
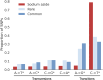
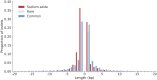
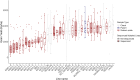
The nature and effect of mutations are of fundamental importance to the evolutionary process. The generation of mutations with mutagens has also played important roles in genetics. Applications of mutagens include dissecting the genetic basis of trait variation, inducing desirable traits in crops, and understanding the nature of genetic load. Previous studies of sodium azide-induced mutations have reported single nucleotide variants (SNVs) found in individual genes. To characterize the nature of mutations induced by sodium azide, we analyze whole-genome sequencing (WGS) of 11 barley lines derived from sodium azide mutagenesis, where all lines were selected for diminution of plant fitness owing to induced mutations. We contrast observed mutagen-induced variants with those found in standing variation in WGS of 13 barley landraces. Here, we report indels that are two orders of magnitude more abundant than expected based on nominal mutation rates. We found induced SNVs are very specific, with C → T changes occurring in a context followed by another C on the same strand (or the reverse complement). The codons most affected by the mutagen include the sodium azide-specific CC motif (or the reverse complement), resulting in a handful of amino acid changes and few stop codons. The specific nature of induced mutations suggests that mutagens could be chosen based on experimental goals. Sodium azide would not be ideal for gene knockouts but will create many missense mutations with more subtle effects on protein function.
Copyright: This is an open access article, free of all copyright, and may be freely reproduced, distributed, transmitted, modified, built upon, or otherwise used by anyone for any lawful purpose. The work is made available under the Creative Commons CC0 public domain dedication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

