Enhanced mutanome analysis towards the induction of neoepitope-reactive T-cell responses for personalized immunotherapy of pancreatic cancer
- PMID: 40461160
- PMCID: PMC12142143
- DOI: 10.1136/jitc-2025-011802
Enhanced mutanome analysis towards the induction of neoepitope-reactive T-cell responses for personalized immunotherapy of pancreatic cancer
Abstract
Background: Personalized immunotherapy of pancreatic ductal adenocarcinoma (PDAC) through T-cell mediated targeting of tumor-specific, mutanome-encoded neoepitopes may offer new opportunities to combat this disease, in particular by countering recurrence after primary tumor resection. However, the sensitive and accurate calling of somatic mutations in PDAC tissue samples is compromised by the low tumor cell content. Moreover, the repertoire of immunogenic neoepitopes in PDAC is limited due to the low mutational load of the majority of these tumors.
Methods: We developed a workflow involving the combined analysis of next-generation DNA and RNA sequencing data from matched pairs of primary tumor samples and patient-derived xenograft models towards the enhanced detection of driver mutations as well as single nucleotide variants encoding potentially immunogenic T-cell neoepitopes. Subsequently, we immunized HLA/human T-cell receptor (TCR) locus-transgenic mice with synthetic peptides representing candidate neoepitopes, and molecularly cloned the genes encoding TCRs targeting these epitopes.
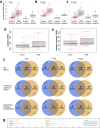
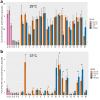
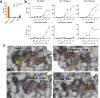
Result: Application of our pipeline resulted in the identification of greater numbers of non-synonymous mutations encoding candidate neoepitopes with increased confidence. Furthermore, we provide proof of concept for the successful isolation of HLA-restricted TCRs from humanized mice immunized with different neoepitopes, several of which would not have been selected based on mutanome analysis of PDAC tissue samples alone. These TCRs mediate specific T-cell reactivity against the tumor cells in which the corresponding mutations were identified.
Conclusion: Enhanced mutanome analysis and candidate neoepitope selection increase the likelihood of identifying therapeutically relevant neoepitopes, and thereby support the optimization of personalized immunotherapy for PDAC and other poorly immunogenic cancers.
Keywords: Adoptive cell therapy - ACT; Gastrointestinal Cancer; Immunotherapy; T cell Receptor - TCR; Vaccine.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: None declared.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
