Down-regulation of neuroprotective protein kinase D in Huntington´s disease
- PMID: 40461488
- PMCID: PMC12134097
- DOI: 10.1038/s41419-025-07688-9
Down-regulation of neuroprotective protein kinase D in Huntington´s disease
Abstract
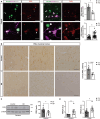
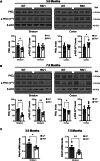
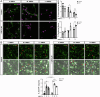
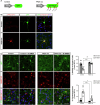
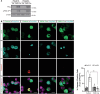
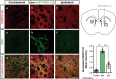
Huntington's disease (HD) is a progressive, autosomal dominant neurodegenerative disorder characterized by the selective dysfunction and loss of neurons in the striatum and cerebral cortex. Experimental evidence suggests that GABAergic medium-sized spiny neurons (MSNs) in the striatum are particularly vulnerable to glutamate-induced toxicity (excitotoxicity) and its analogues. However, the molecular mechanisms underlying MSN-specific death in HD remain poorly understood. The serine/threonine protein kinase D1 (PKD1) confers neuroprotection in various neuropathological conditions, including ischemic stroke. While excitotoxicity inactivates PKD1 in cortical glutamatergic neurons without altering its levels, active PKD1 potentiates the survival of excitatory neurons in highly excitotoxic environments. Here, we investigated whether PKD1 activity dysregulation contributes to MSN death in HD and its association with neurodegeneration. We found an unexpected reduction in PKD1 protein levels in striatal neurons from HD patients. Similarly, the R6/1 mouse model of HD exhibited progressive PKD1 protein loss, commencing at early disease stages, accompanied by decreased Prkd1 transcript levels. PKD1 downregulation also occurred in the cerebral cortex of R6/1 mice, but only at late stages. Functionally, pharmacological PKD inhibition in primary striatal neurons exacerbated excitotoxic damage and apoptosis induced by glutamate N-methyl D-aspartate (NMDA) receptors, whereas expression of constitutively active PKD1 (PKD1-Ca) conferred neuroprotection. Furthermore, PKD1-Ca protected against polyQ-induced apoptosis in a cellular model of HD. In a translational approach, intrastriatal lentiviral delivery of PKD1-Ca in symptomatic R6/1 mice prevented the loss of DARPP-32, a molecular marker of MSNs. Collectively, our findings strongly suggest that PKD1 loss-of-function contributes to HD pathogenesis and the selective vulnerability of MSNs. These findings position PKD1 as a promising therapeutic target for mitigating MSN death in HD.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: The protocol for animal experiments was approved by local ethics committees (IIBM and CSIC) and the Environmental Counseling of the Comunidad de Madrid, Spain. For human samples, a written informed consent for brain removal after death for diagnostic and research purposes was obtained from the brain donors and/or next of kin. Procedures, information and consent forms were approved by the Bioethics Subcommittee of Fundación Cien Madrid, Spain (S19001).
Figures


References
-
- Martin JB, Gusella JF. Huntington’s disease. Pathogenesis and management. N Engl J Med. 1986;315:1267–76. - PubMed
-
- Group THsDCR.. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–83. - PubMed
-
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–84. - PubMed
-
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP Jr. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–77. - PubMed
-
- Graveland GA, DiFiglia M. The frequency and distribution of medium-sized neurons with indented nuclei in the primate and rodent neostriatum. Brain Res. 1985;327:307–11. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

