Modular Control of Boolean Network Models
- PMID: 40461704
- PMCID: PMC12133937
- DOI: 10.1007/s11538-025-01471-9
Modular Control of Boolean Network Models
Abstract
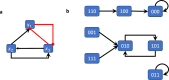
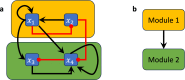
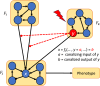
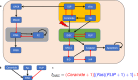
The concept of control is crucial for effectively understanding and applying biological network models. Key structural features relate to control functions through gene regulation, signaling, or metabolic mechanisms, and computational models need to encode these. Applications often focus on model-based control, such as in biomedicine or metabolic engineering. In a recent paper, the authors developed a theoretical framework of modularity in Boolean networks, which led to a canonical semidirect product decomposition of these systems. In this paper, we present an approach to model-based control that exploits this modular structure, as well as the canalizing features of the regulatory mechanisms. We show how to identify control strategies from the individual modules, and we present a criterion based on canalizing features of the regulatory rules to identify modules that do not contribute to network control and can be excluded. For even moderately sized networks, finding global control inputs is computationally challenging. Our modular approach leads to an efficient approach to solving this problem. We apply it to a published Boolean network model of blood cancer large granular lymphocyte (T-LGL) leukemia to identify a minimal control set that achieves a desired control objective.
Keywords: Boolean networks; Canalization; Control; Gene regulatory networks; Modularity.
© 2025. The Author(s).
Figures




Update of
-
Modular control of Boolean network models.ArXiv [Preprint]. 2024 Nov 4:arXiv:2401.12477v3. ArXiv. 2024. Update in: Bull Math Biol. 2025 Jun 3;87(7):91. doi: 10.1007/s11538-025-01471-9. PMID: 38344220 Free PMC article. Updated. Preprint.
Similar articles
-
Modular control of Boolean network models.ArXiv [Preprint]. 2024 Nov 4:arXiv:2401.12477v3. ArXiv. 2024. Update in: Bull Math Biol. 2025 Jun 3;87(7):91. doi: 10.1007/s11538-025-01471-9. PMID: 38344220 Free PMC article. Updated. Preprint.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
An open problem: Why are motif-avoidant attractors so rare in asynchronous Boolean networks?J Math Biol. 2025 Jun 12;91(1):11. doi: 10.1007/s00285-025-02235-8. J Math Biol. 2025. PMID: 40504255 Free PMC article.
-
The Lived Experience of Autistic Adults in Employment: A Systematic Search and Synthesis.Autism Adulthood. 2024 Dec 2;6(4):495-509. doi: 10.1089/aut.2022.0114. eCollection 2024 Dec. Autism Adulthood. 2024. PMID: 40018061 Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Veliz-Cuba A, Arthur J, Hochstetler L, Klomps V, Korpi E (2012) On the relationship of steady states of continuous and discrete models arising from biology. Bull Math Biol 74:2779–2792 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

