Flexible, production-scale, human whole genome sequencing on a benchtop sequencer
- PMID: 40461965
- PMCID: PMC12135483
- DOI: 10.1186/s12864-025-11741-4
Flexible, production-scale, human whole genome sequencing on a benchtop sequencer
Abstract
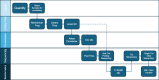
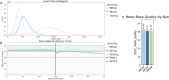
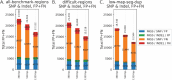
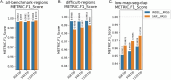
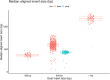
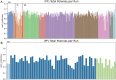
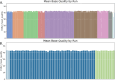
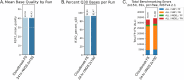
Human whole-genome sequencing (hWGS) provides comprehensive genomic information that can help guide research in disease prevention and treatment. Recent advancements in sequencing technology have improved sequencing quality and further reduced sequencing costs on bench-top sized instruments, making whole-genome sequencing an accessible technology for broader use. Here, we demonstrate the feasibility of a large WGS project using a benchtop sequencer in a small laboratory setting, on a scale previously reserved for production-scale machines. In this project, 807 samples were prepared and sequenced across 313 flow cells, with high sequencing quality at a median %Q30 of 96.6% and a median %Q40 of 89.31%. To screen library quality and maximize sample yield, we utilized 48-plex sample pre-pool 'QC' runs to provide > 1 × coverage per sample prior to sample pooling and full-depth sequencing, providing valuable sample-level insights prior to full-depth sequencing. With this strategy, we consistently achieved > 30 × human whole genome sequencing of three-plex sample trios with standard settings. To demonstrate additional flexibility present in the platform, we explored two different use cases 1) large insert sizes (1kb +) library to achieve superior genome coverage; 2) proof of concept rapid WGS sequencing to minimize sample to answer turnaround time for time-critical sequencing applications. Sequencing of a 2 × 100 > 30 × human WGS can be achieved in < 12 h and subsequent file generation in < 1 additional hour. This study provides a cost-effective and flexible real-world demonstration of achieving both high quality hWGS sequencing and instrument flexibility without the need for complex batching schemes or factory-sized sequencers.
Keywords: Bench Top Sequencer; Benchmarking; Genomics; High-Throughput Sequencing; Human Whole Genome Sequencing; Rapid Whole Genome Sequencing; Sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not Applicable. Consent for publication: All authors have consented to this publication. Competing interests: Competing interests: All authors are current or former employees of Element Biosciences, which has commercialized the sequencing technology described in this paper. They may own stock options in the company.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources

