Associations of plasma protein levels with risk of colorectal cancer: a proteome-wide Mendelian randomization study
- PMID: 40462010
- PMCID: PMC12135285
- DOI: 10.1186/s12014-025-09545-5
Associations of plasma protein levels with risk of colorectal cancer: a proteome-wide Mendelian randomization study
Abstract
Background: The treatment of advanced or metastatic colorectal cancer (CRC) poses a global challenge. Mendelian Randomization (MR) has been primarily applied for repurposing licensed drugs and uncovering new therapeutic targets.
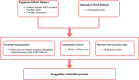
Objective: This study aims to systematically identify potential plasma protein targets for CRC using proteome-wide Mendelian randomization and evaluate their potential side effects through phenome-wide association studies (Phe-WAS).
Methods: We conducted a comprehensive proteome-wide MR study to assess the causal relationships between plasma proteins and the risk of CRC and evaluate their potential side effects through Phe-WAS. The plasma proteins were sourced from the Finland and Iceland decode database, encompassing GWAS data for plasma proteins (Olink-619 samples across 2925 proteins, SomaScan -828 samples across 7596 proteins and Iceland decode database across 4907 proteins). Additionally, GWAS data for CRC were extracted from the UK Biobank-SAIGE database, including 3051 cases and 382,756 controls. Subsequently, colocalization analysis was performed to identify shared causal variants between plasma proteins and CRC. Finally, a phenome-wide association study (Phe-WAS) was conducted to examine the potential adverse effects of druggable proteins for CRC, utilizing the extensive UK Biobank-SAIGE database, encompassing 783 phenotypes.
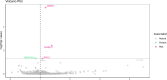
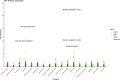
Results: The MR analysis identified GREM1, DKKL1, and CHRDL2 as plasma proteins whose genetically predicted levels were positively associated with CRC risk, whereas TMEM132A was inversely associated with CRC risk (P_fdr < 0.05). The colocalization analysis identified these four proteins as shared variation with CRC (PPH3 + PPH4 > 0.7), suggesting that these proteins represent potential direct targets for CRC intervention. Further phenotype-wide association studies showed no significant potential side effects of these targets (P_fdr > 0.05).
Conclusion: This proteome-wide Mendelian randomization study offers a comprehensive molecular landscape of CRC, identifying GREM1, DKKL1, CHRDL2, and TMEM132A as potential therapeutic targets. Our research provides a critical foundation for future experimental validation and therapeutic development in colorectal cancer management.
Keywords: Colorectal cancer; Drug target; Mendelian randomization; Phenome-wide association study; Plasma proteins.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no competing interests. Ethics approval: This study exclusively utilized publicly available, anonymized genetic summary data from large-scale genomic databases. No individual-level patient data were accessed or processed. Key Ethical Considerations: All source databases obtained appropriate ethical approvals, Data were aggregated and anonymized prior to analysis, No direct human or animal experimentation was conducted, No additional institutional review board (IRB) approval was required, Informed consent for data collection was obtained by the original data repositories. This study adheres to the principles of the Declaration of Helsinki and follows international guidelines for genetic data research. Human and animal rights: No animals/humans were used for studies that are the basis of this research. Consent for publication: Not applicable.
Figures

Similar articles
-
Proteome-wide Mendelian randomization identifies therapeutic targets for ankylosing spondylitis.Front Immunol. 2024 Mar 19;15:1366736. doi: 10.3389/fimmu.2024.1366736. eCollection 2024. Front Immunol. 2024. PMID: 38566994 Free PMC article.
-
Exploring Potential Drug Targets in Multiple Cardiovascular Diseases: A Study Based on Proteome-Wide Mendelian Randomization and Colocalization Analysis.Cardiovasc Ther. 2025 Feb 21;2025:5711316. doi: 10.1155/cdr/5711316. eCollection 2025. Cardiovasc Ther. 2025. PMID: 40026415 Free PMC article.
-
Proteome-wide Mendelian randomization and therapeutic targets for bladder cancer.BMC Urol. 2024 Dec 21;24(1):273. doi: 10.1186/s12894-024-01677-4. BMC Urol. 2024. PMID: 39707285 Free PMC article.
-
PTGES2 and RNASET2 identified as novel potential biomarkers and therapeutic targets for basal cell carcinoma: insights from proteome-wide mendelian randomization, colocalization, and MR-PheWAS analyses.Front Pharmacol. 2024 Jul 5;15:1418560. doi: 10.3389/fphar.2024.1418560. eCollection 2024. Front Pharmacol. 2024. PMID: 39035989 Free PMC article.
-
Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome.Genome Med. 2023 Sep 19;15(1):75. doi: 10.1186/s13073-023-01229-9. Genome Med. 2023. PMID: 37726845 Free PMC article.
References
-
- Morgan E, Arnold M, Gini A, Lorenzoni V, et al. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut. 2023;72:338–44. - PubMed
-
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–91. - PubMed
LinkOut - more resources
Full Text Sources
