Adipocyte/Tumor cell crosstalk via IGF-1/TXNIP axis promotes malignancy and endocrine resistance in breast cancer
- PMID: 40462107
- PMCID: PMC12135404
- DOI: 10.1186/s12964-025-02262-4
Adipocyte/Tumor cell crosstalk via IGF-1/TXNIP axis promotes malignancy and endocrine resistance in breast cancer
Abstract
Background: Despite significant improvements in the outcome of Estrogen Receptor (ER) α-positive breast cancer (BC) following the use of endocrine therapies, resistance remains a major challenge. Clinical studies proved that obesity, in addition to promote BC progression, is associated with a reduced efficacy to these treatments, but mechanisms remain unclear.
Methods: We used co-culture systems followed by validation through an ‘ex vivo’ model of human mammary obese (Ob) adipocytes and obese endocrine-resistant metastatic Patient-Derived Organoids (PDOs). Transcriptomics with MixOmics-MINT and MetaCore Functional Tools along with lentiviral and pharmacological approaches provide insights into mechanisms. Clinical relevance was investigated using public datasets, transcriptome-based (n = 375), and immunohistochemistry-based (n = 65) evaluations.
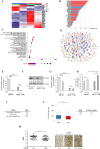
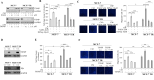
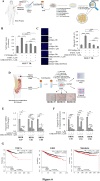
Results: In a model of co-culture, we demonstrated that conditioned media (CM) released by 3T3-L1A adipocytes reduced the sensitivity of parental MCF-7 BC cells to the inhibitory effects of Tamoxifen (Tam) on growth, motility and invasion and significantly increased the proliferative, motile and invasive phenotype of Tam-resistant (TR) BC cells. Transcriptomics identified TXNIP (Thioredoxin-interacting protein), a known tumor suppressor gene, as a network central hub, that was significantly down-regulated in CM-treated MCF-7 and TR cells. Accordingly, TXNIP expression was negatively correlated with Body Mass Index (BMI) in BC patients. Lentiviral TXNIP overexpression and pharmacological induction of TXNIP (i.e. SAHA) or the blockade of insulin-like growth factor-I (IGF-1) signaling, an obesity hallmark able to affect TXNIP expression, reversed CM-mediated effects. TXNIP down-regulation, proliferation and motility in TR cells were exacerbated by CM derived from Ob 3T3-L1A, and combination of an IGF-1 inhibitor and SAHA abrogated Ob-CM activities. Results were also validated in aromatase inhibitor-resistant BC cells. The effectiveness of IGF-1/TXNIP axis inhibition was confirmed using an ‘ex vivo’ model of human mammary obese adipocytes and PDO models. Finally, retrospective analyses demonstrated that an IGF-1high/TXNIPlow signature was correlated with poorer survival in endocrine-treated BC patients.
Conclusions: In conclusion, our study sheds new light on adipocyte/BC cell crosstalk, underscoring the potential of targeting IGF-1/TXNIP axis to block this harmful connection, especially in the context of obesity.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12964-025-02262-4.
Keywords: Adipocytes; Breast Cancer; Endocrine resistance; IGF-1; Obesity; TXNIP; Tumor microenvironment.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was performed in accordance with the Declaration of Helsinki. All clinical specimens and information involved in this study provided informed consent before collection and were approved by The Ethics Committee as specified in Methods’ section. Consent for publication: N/A. Competing interests: The authors declare no competing interests.
Figures



References
-
- Alarcón Rojas CA, Alvarez-Bañuelos MT, Morales-Romero J, Suárez-Díaz H, Hernández-Fonseca JC, Contreras-Alarcón G. Breast cancer: metastasis, molecular subtypes, and overweight and obesity in Veracruz, Mexico. Clin Breast Cancer. 2019;19:e166–71. 10.1016/j.clbc.2018.08.003. - PubMed
Grants and funding
- #2022AA4FTJ/Ministero dell'Istruzione, dell'Università e della Ricerca
- #2017WNKSLR/Ministero dell'Istruzione, dell'Università e della Ricerca
- PE00000019/Ministero dell'Istruzione, dell'Università e della Ricerca
- R01CA072038, BCRF-23-055/BC/NCI NIH HHS/United States
- #26246/Fondazione Italiana per la Ricerca sul Cancro
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

