Gene-Environment Interaction Affects Risk of Atopic Eczema: Population and In Vitro Studies
- PMID: 40462597
- PMCID: PMC12368900
- DOI: 10.1111/all.16605
Gene-Environment Interaction Affects Risk of Atopic Eczema: Population and In Vitro Studies
Erratum in
-
Correction to "Gene-Environment Interaction Affects Risk of Atopic Eczema: Population and In Vitro Studies".Allergy. 2026 Feb;81(2):626. doi: 10.1111/all.70059. Epub 2025 Sep 16. Allergy. 2026. PMID: 40958161 No abstract available.
Abstract
Background: Multiple environmental and genetic factors play a role in the pathogenesis of atopic eczema (AE). We aimed to investigate gene-environment interactions (G × E) to improve understanding of the pathophysiology.
Methods: We analysed data from 16 European studies to test for interaction between the 24 most significant AE-associated loci identified from genome-wide association studies and 18 early-life environmental factors. We tested for replication using a further 10 studies and in vitro modeling to independently assess findings.
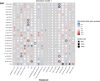
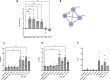
Results: The discovery analysis (including 25,339 individuals) showed suggestive evidence for interaction (p < 0.05) between seven environmental factors (antibiotic use, cat ownership, dog ownership, breastfeeding, elder sibling, smoking and washing practices) and at least one established variant for AE, 14 interactions in total. In the replication analysis (254,532 individuals) dog exposure × rs10214237 (on chromosome 5p13.2 near IL7R) was nominally significant (ORinteraction = 0.91 [0.83-0.99] p = 0.025), with a risk effect of the T allele observed only in those not exposed to dogs. A similar interaction with rs10214237 was observed for siblings in the discovery analysis (ORinteraction = 0.84 [0.75-0.94] p = 0.003), but replication analysis was under-powered (ORinteraction = 1.09 [0.82-1.46]). rs10214237 homozygous risk genotype is associated with lower IL-7R expression in human keratinocytes, and dog exposure modelled in vitro showed a differential response according to rs10214237 genotype.
Conclusion: Interaction analysis and functional assessment provide preliminary evidence that early-life dog exposure may modify the genetic effect of rs10214237 on AE via IL7R, supporting observational epidemiology showing a protective effect for dog ownership. The lack of evidence for other G × E studied here implies only weak effects are likely to occur.
Keywords: atopic eczema; dog; environment; gene; interaction.
© 2025 The Author(s). Allergy published by European Academy of Allergy and Clinical Immunology and John Wiley & Sons Ltd.
Conflict of interest statement
S.J.B. has received research funding (but no personal financial benefits) from the Wellcome Trust (220875/Z/20/Z), UKRI, Medical Research Council, Rosetrees Trust, Stoneygates Trust, British Skin Foundation, Charles Wolfson Charitable Trust, anonymous donations from people with eczema, Unilever, Pfizer, Abbvie, Sosei‐Heptares, Janssen, and European Lead Factory (which includes multiple industry partners). S.J.B., A.B.‐A., K.B., A.D.I., G.H.K., C.S., S.M.L., and L.P. have received funding from the BIOMAP‐IMI consortium (EU H2020 project ref. no. 821511) which receives support from several pharmaceutical industry partners. L.P. has received an honorarium payment for a scientific talk on eczema genetics from LEO Pharma. G.H.K. reports grants from the Netherlands Lung Foundation, ZON‐MW, Ubbo Emmius Foundation, TEVA the Netherlands, GSK, Vertex, outside the submitted work (money to institution). His institution received compensation for consultancy or lectures from Astra Zeneca, Boehringer Ingelheim, and Sanofi.
Figures

Update of
-
Gene-environment interaction analysis in atopic eczema: evidence from large population datasets and modelling in vitro.medRxiv [Preprint]. 2025 Sep 4:2025.01.24.25321071. doi: 10.1101/2025.01.24.25321071. medRxiv. 2025. Update in: Allergy. 2025 Aug;80(8):2201-2212. doi: 10.1111/all.16605. PMID: 40950422 Free PMC article. Updated. Preprint.
References
-
- Johansson S., Bieber T., Dahl R., et al., “Revised Nomenclature for Allergy for Global Use: Report of the Nomenclature Review Committee of the World Allergy Organization, October 2003,” Journal of Allergy and Clinical Immunology 113, no. 5 (2004): 832–836. - PubMed
-
- Mehrmal S., Uppal P., Giesey R. L., and Delost G. R., “Identifying the Prevalence and Disability‐Adjusted Life Years of the Most Common Dermatoses Worldwide,” Journal of the American Academy of Dermatology 82, no. 1 (2020): 258–259. - PubMed
-
- Langan S. M., Irvine A. D., and Weidinger S., “Atopic Dermatitis,” Lancet 396, no. 10247 (2020): 345–360. - PubMed
-
- Odhiambo J. A., Williams H. C., Clayton T. O., Robertson C. F., Asher M. I., and Group IPTS , “Global Variations in Prevalence of Eczema Symptoms in Children From ISAAC Phase Three,” Journal of Allergy and Clinical Immunology 124, no. 6 (2009): 1251–1258.e3. - PubMed
MeSH terms
Grants and funding
- MRC_/Medical Research Council/United Kingdom
- Netherlands Organization for Scientific Research
- Netherlands Organization for Health Research and Development
- ERC_/European Research Council/International
- Netherlands Ministry of Health, Welfare, and Sport
- Lung Foundation Netherlands
- Netherlands Ministry of Spatial Planning, Housing, and the Environment
- Innovative Medicines Initiative
- Asthma and Lung UK
- European Science Foundation
- The North West Lung Centre Charity
- German Federal Ministry of Education and Research
- Ministerio de Ciencia e Innovación
- National Institute for Health and Care Research
- WT_/Wellcome Trust/United Kingdom
- BMA James Trust
- The JP Moulton Charitable Foundation
LinkOut - more resources
Full Text Sources
Miscellaneous

