Analysis of a pan-cancer panel reveals the amino acid metabolism-related gene MTHFD1 as a potential prognostic and immunotherapeutic biomarker
- PMID: 40462856
- PMCID: PMC12131308
- DOI: 10.3892/etm.2025.12892
Analysis of a pan-cancer panel reveals the amino acid metabolism-related gene MTHFD1 as a potential prognostic and immunotherapeutic biomarker
Abstract
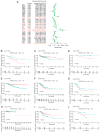
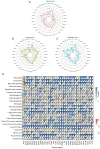
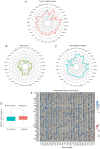
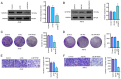
Methylenetetrahydrofolate dehydrogenase 1 (MTHFD1) serves a role in amino acid metabolism and may influence tumor progression. However, to the best of our knowledge, a comprehensive analysis of MTHFD1 in various types of cancer has not been previously performed. Therefore, the present study aimed to investigate the expression profile and prognostic implication of MTHFD1 across various types of cancer, whilst assessing its potential as a novel biomarker and therapeutic target. The expression of MTHFD1 in tissues from various types of cancer was analyzed using online tools based on data from the Cancer Cell Line Encyclopedia and Clinical Proteomic Tumor Analysis Consortium, as well as in-house differential expression analysis using data from The Cancer Genome Atlas (TCGA). The association between MTHFD1 and prognosis was investigated using Kaplan-Meier survival analysis and Cox proportional hazards regression analysis based on TCGA datasets. Furthermore, the association between MTHFD1 and the tumor microenvironment (TME) was investigated using the 'estimation of stromal and immune cells in malignant tumor tissues using expression data' and 'cell-type identification by estimating relative subsets of RNA transcripts' algorithms. The correlation between MTHFD1 expression and tumor mutational burden (TMB), microsatellite instability (MSI) or 48 immune checkpoint blockade-related gene expression levels was investigated using Pearson correlation analyses. The predictive potential of MTHFD1 for immunotherapy efficacy was evaluated using the tumor immune dysfunction and exclusion (TIDE) algorithm with the IMvigor210 dataset. Subsequently, the effects of MTHFD1 on the proliferation and invasion of A549 and 786-O cell lines were assessed using colony formation and Transwell assays. Analysis across 33 tumor types revealed that MTHFD1 expression was significantly upregulated in 12 cancers (e.g., bladder urothelial carcinoma) and downregulated in 6 cancers (e.g., breast invasive carcinoma). Moreover, high MTHFD1 expression was associated with a poorer prognosis in kidney chromophobe and lung adenocarcinoma, but with better prognosis in kidney renal clear cell carcinoma. Additionally, the activity of MTHFD1, evaluated using the single-sample Gene Set Enrichment Analysis algorithm, was significantly upregulated in 21 cancer types, including bladder urothelial carcinoma and breast invasive carcinoma, compared with corresponding normal tissues. MTHFD1expression levels were negatively correlated with immune cell infiltration in 16 tumor types [e.g., adrenocortical carcinoma (ACC)] and positively correlated only in uveal melanoma (UVM). Additionally, MTHFD1 expression levels showed significant correlations with TMB in 17 tumors (e.g., ACC), were negatively correlated with TIDE scores in most tumors except mesothelioma, liver hepatocellular carcinoma, diffuse large B-cell lymphoma and cholangiocarcinoma, and were associated with MSI in 9 tumor types (e.g., UVM). Multivariant Cox regression analysis revealed that MTHFD1 expression was an independent risk factor for prognosis in lung adenocarcinoma, whilst it was an independent protective factor in clear cell renal cell carcinoma, highlighting its distinct prognostic roles in these two tumor types. In vitro experiments found that knocking down or overexpressing MTHFD1 in A549 and 786-O cells, respectively, reduced the corresponding malignant phenotypes. Overall, to the best of our knowledge, results of the present study provided the first comprehensive analysis of MTHFD1 as a potential cancer biomarker and highlighted its role in immune suppression within the TME. These findings suggested that targeting MTHFD1 may be a novel therapeutic strategy, which may enhance the efficacy of immunotherapy and improve the outcomes of patients with various types of cancer.
Keywords: biomarker; immunotherapeutic; methylenetetrahydrofolate dehydrogenase 1; pan-cancer; prognostic.
Copyright: © 2025 Gong et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Identification of SHCBP1 as a potential biomarker involving diagnosis, prognosis, and tumor immune microenvironment across multiple cancers.Comput Struct Biotechnol J. 2022 Jun 18;20:3106-3119. doi: 10.1016/j.csbj.2022.06.039. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35782736 Free PMC article.
-
Pan-Cancer Analysis of PARP1 Alterations as Biomarkers in the Prediction of Immunotherapeutic Effects and the Association of Its Expression Levels and Immunotherapy Signatures.Front Immunol. 2021 Aug 31;12:721030. doi: 10.3389/fimmu.2021.721030. eCollection 2021. Front Immunol. 2021. PMID: 34531868 Free PMC article.
-
Identifies microtubule-binding protein CSPP1 as a novel cancer biomarker associated with ferroptosis and tumor microenvironment.Comput Struct Biotechnol J. 2022 Jun 24;20:3322-3335. doi: 10.1016/j.csbj.2022.06.046. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832625 Free PMC article.
-
[Pan-cancer analysis of ubiquitin-specific protease 7 and its expression changes in the carcinogenesis of scar ulcer].Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2023 Jun 20;39(6):518-526. doi: 10.3760/cma.j.cn501225-20230421-00137. Zhonghua Shao Shang Yu Chuang Mian Xiu Fu Za Zhi. 2023. PMID: 37805766 Free PMC article. Chinese.
-
APOBEC3C is a novel target for the immune treatment of lower-grade gliomas.Neurol Res. 2024 Mar;46(3):227-242. doi: 10.1080/01616412.2023.2287340. Epub 2024 Jan 22. Neurol Res. 2024. PMID: 38007705
References
LinkOut - more resources
Full Text Sources
Miscellaneous
