This is a preprint.
Prolonged non-suppressible viremia sustained by a clonally expanded, genomically defective provirus with an immune-evasive HIV protein expression profile
- PMID: 40462973
- PMCID: PMC12132533
- DOI: 10.1101/2025.05.16.654538
Prolonged non-suppressible viremia sustained by a clonally expanded, genomically defective provirus with an immune-evasive HIV protein expression profile
Abstract
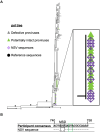
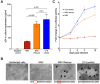
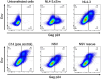
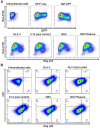
Antiretroviral therapy (ART) potently inhibits HIV replication but does not eliminate HIV proviruses integrated within the genomes of infected cells. Though ART normally suppresses HIV to clinically undetectable levels in blood, some individuals experience non-suppressible viremia (NSV) that is not attributable to suboptimal drug exposure (e.g. due to incomplete medication adherence or pharmacological issues) nor emerging HIV drug resistance, and that does not resolve following regimen modification. We now understand that NSV can originate from expanded cell clones harboring genetically identical proviruses that reactivate en masse to produce clinically detectable viremia. NSV can even originate from proviruses with genomic defects, particularly in HIV's major splice donor (MSD) site, that render the viremia non-infectious. But, because only a limited number of such cases have been described, all in HIV subtype B, the mechanisms that allow cells harboring such proviruses to produce prolonged viremia without being eliminated remain incompletely understood. We characterized a case of MSD-defective, replication-impaired NSV that lasted >4 years in an individual with non-B HIV. Our results reveal that proviruses with MSD deletions can persist by integrating into minimally differentiated CD4+ T-cell subsets that then give rise to the full spectrum of memory and effector subpopulations, and by exhibiting an HIV protein expression profile that would allow these cells to evade cellular and humoral immunity. Our results highlight the need to better understand the biological implications of persistent HIV protein and virion production by genomically defective, clonally expanded proviruses, and for clinical guidelines to acknowledge this type of viremia.
Figures




References
-
- Palich R, Wirden M, Peytavin G, Lê M-P, Seang S, Abdi B, Schneider L, Tubiana R, Valantin M-A, Paccoud O, Soulié C, Calvez V, Katlama C, Marcelin A-G. 2020. Persistent low-level viraemia in antiretroviral treatment-experienced patients is not linked to viral resistance or inadequate drug concentrations. Journal of Antimicrobial Chemotherapy 75:2981–2985. - PubMed
-
- White JA, Wu F, Yasin S, Moskovljevic M, Varriale J, Dragoni F, Camilo-Contreras A, Duan J, Zheng MY, Tadzong NF, Patel HB, Quiambao JMC, Rhodehouse K, Zhang H, Lai J, Beg SA, Delannoy M, Kilcrease C, Hoffmann CJ, Poulin S, Chano F, Tremblay C, Cherian J, Barditch-Crovo P, Chida N, Moore RD, Summers MF, Siliciano RF, Siliciano JD, Simonetti FR. 2023. Clonally expanded HIV-1 proviruses with 5′-leader defects can give rise to nonsuppressible residual viremia. J Clin Invest 133. - PMC - PubMed
-
- Halvas EK, Joseph KW, Brandt LD, Guo S, Sobolewski MD, Jacobs JL, Tumiotto C, Bui JK, Cyktor JC, Keele BF, Morse GD, Bale MJ, Shao W, Kearney MF, Coffin JM, Rausch JW, Wu X, Hughes SH, Mellors JW. 2020. HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. J Clin Invest 130:5847–5857. - PMC - PubMed
-
- Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, Frenkel LM. 2005. Evidence that Low-Level Viremias during Effective Highly Active Antiretroviral Therapy Result from Two Processes: Expression of Archival Virus and Replication of Virus. J Virol 79:9625–9634. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
