This is a preprint.
Serine auxotrophy is a targetable vulnerability driven by PSAT1 suppression in AML
- PMID: 40462988
- PMCID: PMC12132333
- DOI: 10.1101/2025.05.13.651470
Serine auxotrophy is a targetable vulnerability driven by PSAT1 suppression in AML
Abstract
Serine metabolism is of growing biologic and therapeutic interest in cancer. Upregulation of the serine synthesis pathway (SSP) can fuel tumor growth, and cancers with this phenotype are often sensitive to SSP inhibitors. In parallel, dietary restriction of serine and glycine (SG) can suppress some cancers, but the determinants of sensitivity to this approach are poorly understood. This is especially true in acute myeloid leukemia (AML), where serine metabolism has been less explored. We report that a subset of human AML cell lines and primary samples are completely dependent on external serine, known as serine auxotrophy. These leukemias consistently suppressed the SSP enzyme PSAT1, failed to synthesize serine, responded to SG restriction in vivo, and were rescued by restoring PSAT1. We also found that AML with an SF3B1 K700E mutation showed additional dependence on the SSP enzyme PHGDH, that SG restriction synergized with venetoclax in serine auxotrophic AML, and that MECOM rearrangement was strongly associated with PSAT1 suppression and serine auxotrophy. These findings define a metabolically distinct AML subtype and nominate it for targeting by SG restriction.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures


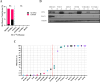



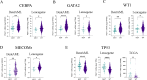

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
