This is a preprint.
A complete map of human cytosolic degrons and their relevance for disease
- PMID: 40463067
- PMCID: PMC12132302
- DOI: 10.1101/2025.05.10.653233
A complete map of human cytosolic degrons and their relevance for disease
Abstract
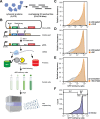
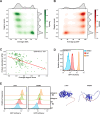
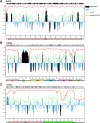
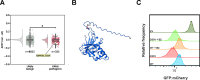
Degrons are short protein segments that target proteins for degradation via the ubiquitin-proteasome system and thus ensure timely removal of signaling proteins and clearance of misfolded proteins from the intracellular space. Here, we describe a systematic screen for degrons in the human cytosol. We determine degron potency of >200,000 different 30-residue tiles from more than 5,000 cytosolic human proteins with 99.7% coverage. In total, 19.1% of the tiles function as strong degrons, 30.4% as intermediate degrons, while 50.5% did not display degron properties. The vast majority of the degrons are dependent on the E1 ubiquitin-activating enzyme and the proteasome but independent of autophagy. The results reveal both known and novel degron motifs, both internal as well as at the C-terminus. Mapping the degrons onto protein structures, predicted by AlphaFold2, revealed that most of the degrons are located in buried regions, indicating that they only become active upon unfolding or misfolding. Training of a machine learning model allowed us to probe the degron properties further and predict the cellular abundance of missense variants that operate by forming degrons in exposed and disordered protein regions, thus providing a mechanism of pathogenicity for germline coding variants at such positions.
Keywords: DMS; E3; chaperone; deep mutational scanning; gene variants; proteasome; protein degradation; protein folding; protein quality control; protein stability; ubiquitin.
Conflict of interest statement
Competing interests K.L-L. holds stock options in and is a consultant for Peptone Ltd. All other authors declare no competing interests.
Figures


References
-
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. - PubMed
-
- Schimke RT, Doyle D. Control of enzyme levels in animal tissues. Annu Rev Biochem. 1970;39:929–76. - PubMed
-
- Eden E, Geva-Zatorsky N, Issaeva I, Cohen A, Dekel E, Danon T, et al. Proteome half-life dynamics in living human cells. Science. 2011;331(6018):764–8. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
