This is a preprint.
Selective GSK3α Inhibition Promotes Self-Renewal Across Different Stem Cell States
- PMID: 40463072
- PMCID: PMC12132347
- DOI: 10.1101/2025.05.16.653860
Selective GSK3α Inhibition Promotes Self-Renewal Across Different Stem Cell States
Abstract
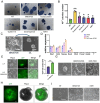
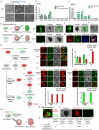
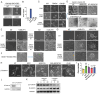
Pan-GSK3α/β inhibition promotes stem cell self-renewal through activation of WNT/β-catenin signaling, but its broad effects complicate the precise control of stem cell states. Here, we show that selective inhibition of GSK3α with BRD0705 supports the long-term self-renewal of mouse embryonic stem cells (ESCs), epiblast stem cells (EpiSCs), and neural stem cells (NSCs), independent of β-catenin signaling. When combined with the tankyrase inhibitor IWR1, BRD0705 broadly supports the maintenance of diverse pluripotent stem cell states, including ESCs, EpiSCs, and formative pluripotent stem cells. This BRD0705/IWR1 cocktail enables stable co-culture of naive ESCs and primed EpiSCs while preserving their distinct molecular and functional identities. Single-cell transcriptomics, epigenomic profiling, and functional assays confirm sustained lineage-specific features across stem cell types. These findings demonstrate that selective GSK3α inhibition enhances stemness by buffering against differentiation cues and promoting intrinsic self-renewal capacity. This work identifies GSK3α as a key regulator of self-renewal across distinct stem cell states and establishes a versatile culture system with broad applications.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests. 1 provisional patent related to this study have been filed (APPLICATION # 63/798,735).
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
