This is a preprint.
Cryo-EM evidence for a common factor in Alzheimer's and other neurodegenerations
- PMID: 40463114
- PMCID: PMC12132381
- DOI: 10.1101/2025.05.13.653829
Cryo-EM evidence for a common factor in Alzheimer's and other neurodegenerations
Abstract
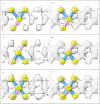
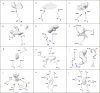
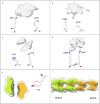
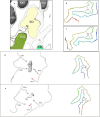
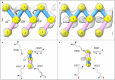
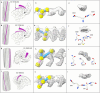
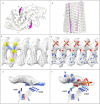
In the last seven years, cryo-EM maps of neuropathological fibrils from Alzheimer's disease and other neurodegenerations have been released by various authors1-44. The first publication11 noted an unknown component coordinating with lysine residues in the protein, a finding recapitulated in many succeeding studies. Previous authors have emphasized difficulties in analysing this component12,20,28,33,43,45, but current findings, using powerful visualisation software UCSF ChimeraX46 on all publicly available maps1-44, indicate that the issue is tractable. Lysine-coordinating extra densities have common features, including a Y-shaped substructure, suggestive of a molecular factor in common, in neuropathological fibrils from a wide range of neurodegenerations and involving misfolded proteins beta-amyloid10,35, alpha-synuclein27,37,39,41, prion protein17, tau1,5,7,8,11,12,15,16,19,22-26,29-33,35,43 and transmembrane protein 106B5,9,18,20,24,28,36,44. A similar component, albeit in non-lysine environments, was found in neuropathological fibrils involving TAR DNA-binding protein 432,3 and TATA-binding protein-associated factor 1536. The results suggest the existence of a common molecular factor, a predominantly anionic polymer, linking these diseases and raising the possibility of a unitary basis for Alzheimer's and other neurodegenerations. Based on evidence here, RNA is a feasible candidate for this putative common factor. Such findings raise the possibility of new diagnostic tests and treatments for these devastating diseases in the future.
Figures





Similar articles
-
RNA as a component of scrapie fibrils.Sci Rep. 2024 Feb 29;14(1):5011. doi: 10.1038/s41598-024-55278-0. Sci Rep. 2024. PMID: 38424114 Free PMC article.
-
Replicating RNA as a component of scrapie fibrils.bioRxiv [Preprint]. 2023 Aug 18:2023.08.17.553578. doi: 10.1101/2023.08.17.553578. bioRxiv. 2023. Update in: Sci Rep. 2024 Feb 29;14(1):5011. doi: 10.1038/s41598-024-55278-0. PMID: 37645951 Free PMC article. Updated. Preprint.
-
High-resolution Cryo-EM Structure Determination of a-Synuclein-A Prototypical Amyloid Fibril.Bio Protoc. 2025 Feb 5;15(3):e5171. doi: 10.21769/BioProtoc.5171. eCollection 2025 Feb 5. Bio Protoc. 2025. PMID: 39959285 Free PMC article.
-
Emerging Trends in Cryo-EM-based Structural Studies of Neuropathological Amyloids.J Mol Biol. 2023 Dec 15;435(24):168361. doi: 10.1016/j.jmb.2023.168361. Epub 2023 Nov 8. J Mol Biol. 2023. PMID: 37949311 Review.
-
Pathogenic mechanisms of prion protein, amyloid-β and α-synuclein misfolding: the prion concept and neurotoxicity of protein oligomers.J Neurochem. 2016 Oct;139(2):162-180. doi: 10.1111/jnc.13772. Epub 2016 Sep 15. J Neurochem. 2016. PMID: 27529376 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
