An integrated mutation-based immunoinformatic approach incorporating variability in epitopes: a study based on HIV subtype C
- PMID: 40463364
- PMCID: PMC12129944
- DOI: 10.3389/fimmu.2025.1540253
An integrated mutation-based immunoinformatic approach incorporating variability in epitopes: a study based on HIV subtype C
Abstract
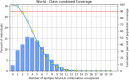
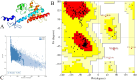
Currently, HIV (human immunodeficiency virus) infection is one of the leading complications in public health and causes acquired immunodeficiency syndrome (AIDS), especially in the African region. No specific vaccine is available to combat this, with multi-strain variability being one of the hurdles. In this investigation, we employed variability in the epitope of the HIV subtype C targets to introduce mutations and construct an epitope-based vaccine. Four targets were examined to predict the B and T cells (major histocompatibility complex class I and II). Among the predicted epitopes, immunodominant epitopes were selected and were mapped with the identified variable amino acid to incorporate mutation. These selected and mutated epitopes were used for the non-mutated and mutated vaccine construction, considering linker for fusion and adjuvant to improve the activity. The vaccine's structure was modeled and examined to validate its structural quality, and a high population coverage was also found. The docking investigation of the non-mutated and mutated vaccine with Toll-like receptor 3 shows remarkable activity followed by strong binding affinity, and the simulation of over 100 ns revealed the constancy of the complex system. The immune response revealed its strong effectiveness by generating multiple immunoglobulins followed by the time step of infection, and further, in silico cloning demonstrated a high expression in Escherichia coli based on their favorable Codon Adaptation Index and GC value. The integrated approach in this investigation will help to plan a potent immunodominant vaccine that can work for multiple strains of HIV infection.
Keywords: HIV; TLR3; epitope; mutation; simulation; vaccine.
Copyright © 2025 Kumar Mishra, Kumar, Or Rashid, Sultana, Dawoud, Bourhia and Georrge.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
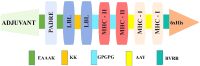




References
-
- Hashempour A, Khodadad N, Akbarinia S, Ghasabi F, Ghasemi Y, Nazar MMKA, et al. Reverse vaccinology approaches to design a potent multiepitope vaccine against the HIV whole genome: immunoinformatic, bioinformatics, and molecular dynamics approaches. BMC Infect Dis. (2024) 24:873. doi: 10.1186/s12879-024-09775-2 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

