Impact of embryo size on apoptosis in C. elegans
- PMID: 40463483
- PMCID: PMC12131072
- DOI: 10.17912/micropub.biology.001608
Impact of embryo size on apoptosis in C. elegans
Abstract
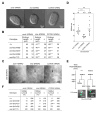
During C. elegans development 131 somatic cells reproducibly undergo programmed cell death. Many of these 131 cells 'programmed to die' are the smaller daughter of a neuroblast that divides asymmetrically and die through apoptosis. To determine whether cell size impacts the ability of cells programmed to die to undergo apoptosis, we increased or decreased embryo size by RNA interference-mediated knock-down of the genes C27D9.1 or ima-3 , respectively. We found that in apoptosis-compromised genetic backgrounds, C27D9.1 ( RNAi ) enhances and ima-3 ( RNAi ) partially suppresses inappropriate survival of cells programmed to die. This supports the notion that in C. elegans embryos, an increase in cell size compromises and a decrease in cell size promotes the ability of cells programmed to die to undergo apoptosis.
Copyright: © 2025 by the authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest present.
Figures
References
-
- Audhya Anjon, Hyndman Francie, McLeod Ian X., Maddox Amy S., Yates John R., Desai Arshad, Oegema Karen. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans . The Journal of Cell Biology. 2005 Oct 24;171(2):267–279. doi: 10.1083/jcb.200506124. - DOI - PMC - PubMed
-
- Benjamini Yoav, Hochberg Yosef. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B: Statistical Methodology. 1995 Jan 1;57(1):289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
LinkOut - more resources
Full Text Sources
