Impact of ITH on PRAD patients and feasibility analysis of the positive correlation gene MYLK2 applied to PRAD treatment
- PMID: 40463716
- PMCID: PMC12130005
- DOI: 10.3389/fgene.2025.1589259
Impact of ITH on PRAD patients and feasibility analysis of the positive correlation gene MYLK2 applied to PRAD treatment
Abstract
Introduction: Prostate adenocarcinoma (PRAD) is an extremely widespread site of urological malignancy and is the second most common male cancer in the world. Currently, research progress in immunotherapy for prostate treatment is slower compared to other tumours, which is mainly considered to be caused by the low rate of immune response in prostate cancer as a cold tumour. Recent studies have shown that intra-tumour heterogeneity (ITH) is an important impediment to PRAD immunotherapy. Therefore, we set out to investigate the feasibility of judging patients' disease and knowing the clinical treatment based on the level of ITH.
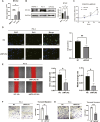
Methods: Clinical information and transcriptome expression matrices of PRAD samples were gained from The Cancer Genome Atlas (TCGA) database. The ITH-score of PRAD samples was evaluated using the DEPTH algorithm. The optimal cut-off value of RiskScore was calculated based on the difference in survival curves, and PRAD patients were classified into high ITH and low ITH groups based on the optimal cut-off value. Genes with expression differences were screened by differential expression gene analyses (DEGs), and 103 positively correlated differentially expressed genes were identified based on these genes as well as the ITH-score. We conducted multivariate Cox regression to sift for prognostically relevant genes to structure an ITH-related prognostic signature. GO and KEGG pathway enrichment analyses were performed on these 103 positively correlated differentially expressed genes, and the proportion and type of tumour-infiltrating immune cells were assessed by TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL and EPIC algorithms in patients. In addition, we calculated the relevance of immunotherapy and predicted various drugs that might be used for treatment and evaluated the predictive power of survival models under multiple machine learning algorithms through the training set TCGA-PRAD versus the validation set PRAD-FR cohort. Based on the upregulated differential gene and ITH-score correlation ranking, combined with the prognostic performance of the gene, we chose MYLK2 as an elite gene for ITH, and performed cellular experiments to validate it by PCR and WB, as well as CCK8, scratch experiments, and transwell experiments on si-MYLK2 PRAD. Finally, we constructed cox regression models as well as random forest survival models based on the expression levels of SYNPO2L, MYLK2, CKM and MYL3.
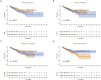
Results: We found that lowering the ITH-score resulted in better survival outcomes. We identified 20 highly correlated differentially expressed genes by calculating the correlation coefficient (cor>0.3) between them by DEGs as well as ITH-score, and selected four genes with p-value less than 0.05 (SYNPO2L, MYLK2, CKM and MYL3) by combining with cox regression. Survival analysis based on the differential expression grouping of SYNPO2L, MYLK2, CKM and MYL3 suggested significant survival differences. The results of biofunctional pathway enrichment analysis suggested that the PRAD-ITH gene set had significant expression in the Mucsle Contraction pathway. Macroscopic differences in the immune landscape and differences in responsiveness to immunotherapy existed between ITH-H and ITH-L. The results of the CMap data suggested that NU.1025 was the most likely drug to treat PRAD. The results of our machine learning model constructed based on ITH-score suggest that the random survival forest (RSF) model performs well in both the training and validation sets and has the potential to be used as a clinical prediction model. In vitro experiments verified that MYLK2 plays an important role in the proliferation and migration of PRAD. Our results suggest that the implementation of therapeutic strategies based on key ITH genes may bring new hope for PRAD patients.
Discussion: Our findings indicate that ITH may be an important biomarker for the prognosis and characterisation of PRAD and that the ITH-related gene MYLK2 may serve as a novel target for the treatment of PRAD patients.
Keywords: MYLK2; immunotherapy; intra-tumour heterogeneity (ITH); prognosis; prostate adenocarcinoma (PRAD).
Copyright © 2025 Ma, Li, Song, Qi and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

