Harvard Electroencephalography Database: A comprehensive clinical electroencephalographic resource from four Boston hospitals
- PMID: 40464151
- PMCID: PMC12455399
- DOI: 10.1111/epi.18487
Harvard Electroencephalography Database: A comprehensive clinical electroencephalographic resource from four Boston hospitals
Abstract
Objective: This article presents the Harvard Electroencephalography Database (HEEDB), a large-scale, deidentified, and standardized electroencephalographic (EEG) resource supporting artificial intelligence-driven and reproducible research in epilepsy and broader clinical neuroscience.
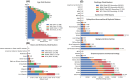
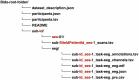
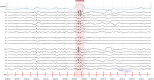
Methods: HEEDB aggregates more than 280 000 EEG recordings from more than 108 000 patients across four Harvard-affiliated hospitals. Data are harmonized using the Brain Imaging Data Structure and hosted on the Brain Data Science Platform. EEG data are linked with clinical notes, International Classification of Diseases, 10th Revision codes, medications, and EEG reports. Deidentification follows Health Insurance Portability and Accountability Act Safe Harbor standards.
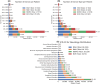
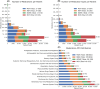
Results: The database includes routine, epilepsy monitoring unit, and intensive care unit EEGs across all age groups, with 73% linked to deidentified clinical reports and 96% of those matched to recordings. Findings are extracted using expert curation, regular expressions, and medical natural language processing models. Auxiliary data include diagnoses, medications, and hospital course, supporting multimodal analysis.
Significance: HEEDB fills a critical gap in EEG data availability for epilepsy research. By enabling large-scale, privacy-compliant, and clinically relevant analysis, it accelerates the development of diagnostic tools, improves training datasets for machine learning, and promotes data-sharing in alignment with FAIR (Findable, Accessible, Interoperable, Reusable) and National Institutes of Health data policies.
Keywords: AI for neurology; Data‐driven EEG analysis; Deidentified clinical data; EEG data platform; EEG large‐scale database.
© 2025 The Author(s). Epilepsia published by Wiley Periodicals LLC on behalf of International League Against Epilepsy.
Conflict of interest statement
M.B.W. is a cofounder of and consultant to Beacon Biosignals, with personal equity, and receives royalties from Wolters Kluwer and Demos Medical. D.M.G. is an unpaid advisor for Epilepsy AI and Eysz, and a paid advisor for Magic Leap. He has received speaker fees from AAN, AES, ACNS, NNS, and AI in Epilepsy and Neurology, and served as a consultant for Neuro Event Labs, IDR, LivaNova, and Health Advances. T.L. is an inventor on patents and patent applications related to the detection, prediction, management, and treatment of epilepsy and seizures; has received device donations from Epitel and Empatica; has received travel support from academic and scientific organizations; and hosts international fellows. C.T.S. and B.G. are employed by and hold equity in Amazon Web Services. J.R. is the founder of the Global Brain Care Coalition and cofounder of McCance for Brain Health, has consulted for the NFL and Eli Lilly, and holds leadership roles at Columbia University,
Figures

References
-
- NOT‐OD‐21‐013: Final NIH Policy for Data Management and Sharing. Available from: https://grants.nih.gov/grants/guide/notice‐files/NOT‐OD‐21‐013.html
-
- Cloud Life Sciences public datasets | Cloud Life Sciences Documentation. Google Cloud. Available from: https://cloud.google.com/life‐sciences/docs/resources/public‐datasets
-
- Open Data Sponsorship Program | AWS. Amazon Web Services, Inc. Available from: https://aws.amazon.com/opendata/open‐data‐sponsorship‐program/.
MeSH terms
Grants and funding
- W81XWH-19-1-0861/U.S. Department of Defense
- Zoll Foundation
- RF1AG064312/NH/NIH HHS/United States
- R01HL161253/NH/NIH HHS/United States
- R01 NS131347/NS/NINDS NIH HHS/United States
- R01 AG073410/AG/NIA NIH HHS/United States
- 1OT2OD032701/NH/NIH HHS/United States
- R01 NS128342/NS/NINDS NIH HHS/United States
- R01NS128342/NH/NIH HHS/United States
- Hellman Foundation
- K23NS119794/NH/NIH HHS/United States
- 2022CAL-Amorim/Cures Within Reach
- RF1NS120947/NH/NIH HHS/United States
- R01AG073410/NH/NIH HHS/United States
- R01 EB032820/EB/NIBIB NIH HHS/United States
- R01NS130119/NH/NIH HHS/United States
- RF1 NS120947/NS/NINDS NIH HHS/United States
- Beth Israel Deaconess Medical Center
- R01NS126282/NH/NIH HHS/United States
- 24DIVSUP1274116/American Heart Association
- R01 HL161253/HL/NHLBI NIH HHS/United States
- K23 NS124656/NS/NINDS NIH HHS/United States
- ERP 220036/U.S. Department of Defense
- K23NS124656/NH/NIH HHS/United States
- RF1 AG064312/AG/NIA NIH HHS/United States
- R01NS131347/NH/NIH HHS/United States
- OT2 OD032701/OD/NIH HHS/United States
- R01 NS130119/NS/NINDS NIH HHS/United States
- 20CDA35310297/American Heart Association
- R01AG073598/NH/NIH HHS/United States
- R01 AG073598/AG/NIA NIH HHS/United States
- 2014431/National Science Foundation
- K23 NS119794/NS/NINDS NIH HHS/United States
- K99NS138680/NH/NIH HHS/United States
- K99 NS138680/NS/NINDS NIH HHS/United States
- W81XWH-21-C-0075/U.S. Department of Defense
- R01 NS126282/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

