Uncovering the Unusual Inhibition Mechanism of a Trypanosome Alternative Oxidase Inhibitor Displaying Broad-Spectrum Activity against African Animal Trypanosomes
- PMID: 40464345
- PMCID: PMC12406258
- DOI: 10.1021/acs.jmedchem.5c00631
Uncovering the Unusual Inhibition Mechanism of a Trypanosome Alternative Oxidase Inhibitor Displaying Broad-Spectrum Activity against African Animal Trypanosomes
Abstract
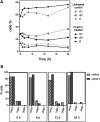
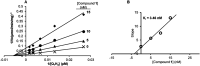
The glucose-dependent respiration of bloodstream forms of the parasite Trypanosoma brucei depends on an unusual and essential mitochondrial electron-transport system, consisting of glycerol-3-phosphate dehydrogenase and the trypanosome alternative oxidase (TAO). We report here the discovery of an allosteric inhibitor of TAO that displays highly potent activity (EC50 values in the range 1-20 nM) against the important veterinary pathogens T. b. brucei, Trypanosoma evansi, Trypanosoma equiperdum, and Trypanosoma congolense, i.e., >5-fold greater potency than the standard drugs. The methylene-linked 2-methyl-4-hydroxybenzoate 2-pyridinyldiphenylphosphonium derivative (1) was the best inhibitor of recombinant TAO (IC50 = 1.3 nM) via a noncompetitive/allosteric mechanism (Ki = 3.46 nM). Remarkably, X-ray crystallography showed that 1 was bound to a site of TAO ∼25 Å from the catalytic pocket. Although 1 demonstrated good safety toward mammalian cells in vitro (selectivity index >2300), it did not fully clear parasitemia in experimental animals, attributable to a high hepatic clearance.
Figures
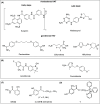




 , while the triphenylphosphonium tail as
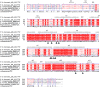
☆, both linked by the methylene linker. The electron density
map is shown contoured to 1.0 σ. (C) The amino acids binding
compound 1. The amino acid residues located within 4
Å of the 1 molecule are shown in stick models. The
hydrogen bonds are shown in dash lines. The omit electron density
map for molecule 1 is contoured 1.0 σ (blue) and
1.5 σ (purple), respectively. (D) Dimer structure of TAO–1 complex. Chain A is shown in the rainbow cartoon from blue
(N terminus) to red (C terminus), and chain B is gray. Compound 1 is shown in the yellow stick model. Fe atoms and OH– ions are shown as purple and red spheres, respectively.
Compound 1 is bound only to chain A, while the Fe atoms
and OH– ion are in both chains. (E) 90° orientation
of chain D, revealing that 1 was bound far away (∼25
Å) from the active site of TAO. Fe atoms and OH– ions present in the active site are shown as purple and red spheres,
respectively.
, while the triphenylphosphonium tail as
☆, both linked by the methylene linker. The electron density
map is shown contoured to 1.0 σ. (C) The amino acids binding
compound 1. The amino acid residues located within 4
Å of the 1 molecule are shown in stick models. The
hydrogen bonds are shown in dash lines. The omit electron density
map for molecule 1 is contoured 1.0 σ (blue) and
1.5 σ (purple), respectively. (D) Dimer structure of TAO–1 complex. Chain A is shown in the rainbow cartoon from blue
(N terminus) to red (C terminus), and chain B is gray. Compound 1 is shown in the yellow stick model. Fe atoms and OH– ions are shown as purple and red spheres, respectively.
Compound 1 is bound only to chain A, while the Fe atoms
and OH– ion are in both chains. (E) 90° orientation
of chain D, revealing that 1 was bound far away (∼25
Å) from the active site of TAO. Fe atoms and OH– ions present in the active site are shown as purple and red spheres,
respectively.

Similar articles
-
Inhibition of trypanosome alternative oxidase without its N-terminal mitochondrial targeting signal (ΔMTS-TAO) by cationic and non-cationic 4-hydroxybenzoate and 4-alkoxybenzaldehyde derivatives active against T. brucei and T. congolense.Eur J Med Chem. 2018 Apr 25;150:385-402. doi: 10.1016/j.ejmech.2018.02.075. Epub 2018 Feb 26. Eur J Med Chem. 2018. PMID: 29544150
-
African trypanosomiasis: Synthesis & SAR enabling novel drug discovery of ubiquinol mimics for trypanosome alternative oxidase.Eur J Med Chem. 2017 Dec 1;141:676-689. doi: 10.1016/j.ejmech.2017.09.067. Epub 2017 Oct 6. Eur J Med Chem. 2017. PMID: 29107420 Free PMC article.
-
Synthesis, biological, and photophysical studies of molecular rotor-based fluorescent inhibitors of the trypanosome alternative oxidase.Eur J Med Chem. 2021 Aug 5;220:113470. doi: 10.1016/j.ejmech.2021.113470. Epub 2021 Apr 16. Eur J Med Chem. 2021. PMID: 33940464
-
Trypanosome alternative oxidase: from molecule to function.Trends Parasitol. 2006 Oct;22(10):484-91. doi: 10.1016/j.pt.2006.08.007. Epub 2006 Aug 21. Trends Parasitol. 2006. PMID: 16920028 Review.
-
The trypanosome alternative oxidase: a potential drug target?Parasitology. 2018 Feb;145(2):175-183. doi: 10.1017/S0031182016002109. Epub 2016 Nov 29. Parasitology. 2018. PMID: 27894362 Review.
References
-
- Franco J. R., Cecchi G., Paone M., Diarra A., Grout L., Kadima Ebeja A., Simarro P. P., Zhao W., Argaw D.. The elimination of human African trypanosomiasis: Achievements in relation to WHO road map targets for 2020. PLoS Neglected Trop. Dis. 2022;16(1):e0010047. doi: 10.1371/journal.pntd.0010047. - DOI - PMC - PubMed
-
- Franco J. R., Cecchi G., Priotto G., Paone M., Diarra A., Grout L., Simarro P. P., Zhao W., Argaw D.. Monitoring the elimination of human African trypanosomiasis at continental and country level: Update to 2018. PLoS Neglected Trop. Dis. 2020;14(5):e0008261. doi: 10.1371/journal.pntd.0008261. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

