A group of segmented viruses contains genome segments sharing homology with multiple viral taxa
- PMID: 40464578
- PMCID: PMC12282112
- DOI: 10.1128/jvi.00332-25
A group of segmented viruses contains genome segments sharing homology with multiple viral taxa
Abstract
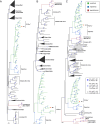
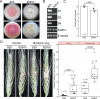
The discovery of diverse segmented RNA viruses through metatranscriptomics has enabled researchers to trace their evolutionary trajectories. However, this effort has been hindered by the limited availability of complete genome sequences and the low similarity of novel viral segments. In this study, we characterized Fusarium asiaticum vivivirus 1 (FaVvV1), a +ssRNA mycovirus with 10 monocistronic RNA segments (S1 to S10, encoding VP1 to VP10), present in the phytopathogenic fungus Fusarium asiaticum. VP1 and VP2 exhibit homology with replication proteins of martellivirals, while VP3 and VP5 share similarities with the nuclear inclusion protein a and the cylindrical inclusion helicase of potyvirids, respectively. FaVvV1 forms rod-shaped virions, with VP8 functioning as a structural protein resembling the helical capsid of potyvirids and closterovirids. To explore the conservation and evolution of viviviruses, we mined 23 public Sequence Read Archive (SRA) datasets, identifying 29 vivivirus-related viruses (vivivirids) comprising 186 viral segments. VP1 (methyltransferase and RdRP domain), VP2 (methyltransferase and superfamily 1 helicase domain), VP3 (chymotrypsin-type serine protease domain), VP5 (superfamily 2 helicase domain), and VP8 (helical capsid) were identified as conserved hallmark proteins of viviviruses. Phylogenetic and structural analyses suggest that multiple genome segmentations and gene/domain duplications were involved in the evolution of vivivirids. VP3, VP5, and VP8 might share a common ancestor with potyvirids. These findings highlight the intricate evolutionary mechanisms underlying segmented virus diversity and adaptation.
Importance: Metaviromics has greatly expanded our understanding of viral diversity, including segmented or multipartite RNA viruses with genomes composed of multiple segments. However, virome analyses often fail to detect genomic segments beyond the RdRP, likely due to their low similarity to known viruses. We characterized a group of segmented, potentially multipartite, +ssRNA viruses, with Fusarium asiaticum vivivirus 1 as a representative; most of these viruses likely infect fungi. Through structural and evolutionary analysis of the five core segments of viviviruses, our findings highlight key aspects of vivivirus evolution, including genome segmentation, gene and domain duplications, and segments with multiple evolutionary origins.
Keywords: genome segmentation; mycovirus; rod-shaped virion; segmented virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Characterization of a multi-segmented rod-shaped mycovirus within the order Martellivirales largely accommodating plant viruses.Virus Res. 2025 Jul;357:199591. doi: 10.1016/j.virusres.2025.199591. Epub 2025 May 30. Virus Res. 2025. PMID: 40451551 Free PMC article.
-
Novel polymycoviruses are encapsidated in filamentous virions.J Virol. 2025 Jan 31;99(1):e0151524. doi: 10.1128/jvi.01515-24. Epub 2024 Dec 10. J Virol. 2025. PMID: 39655956 Free PMC article.
-
A novel tobamo-like mycovirus with filamentous particles replicates in plant cells.J Virol. 2025 May 20;99(5):e0210224. doi: 10.1128/jvi.02102-24. Epub 2025 Mar 31. J Virol. 2025. PMID: 40162784 Free PMC article.
-
How lived experiences of illness trajectories, burdens of treatment, and social inequalities shape service user and caregiver participation in health and social care: a theory-informed qualitative evidence synthesis.Health Soc Care Deliv Res. 2025 Jun;13(24):1-120. doi: 10.3310/HGTQ8159. Health Soc Care Deliv Res. 2025. PMID: 40548558
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
Cited by
-
Characterization of a multi-segmented rod-shaped mycovirus within the order Martellivirales largely accommodating plant viruses.Virus Res. 2025 Jul;357:199591. doi: 10.1016/j.virusres.2025.199591. Epub 2025 May 30. Virus Res. 2025. PMID: 40451551 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

