Serum metabolomics reveals systemic metabolic alterations of Graves' disease before and after antithyroid drug treatment
- PMID: 40465360
- PMCID: PMC12177881
- DOI: 10.1530/EC-25-0078
Serum metabolomics reveals systemic metabolic alterations of Graves' disease before and after antithyroid drug treatment
Abstract
Objective: This study aimed to utilize untargeted metabolomics to analyze changes of serum metabolites before and after methimazole treatment in patients with Graves' disease (GD).
Methods: Total 40 GD patients and 30 healthy volunteers were recruited for the study. Metabolomics analysis was conducted using liquid chromatography-mass spectrometry.
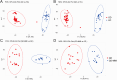
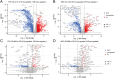
Results: A total of 11,590 metabolites were measured. Compared to healthy controls, newly diagnosed untreated GD patients exhibited significant metabolic dysregulation, with 1,805 metabolites upregulated and 1,737 downregulated. After 1 year of methimazole treatment and normalization of thyroid hormone levels, 137 metabolites remained upregulated and 242 remained downregulated, suggesting incomplete metabolic recovery. Pathway enrichment analysis indicated significant alterations in tyrosine metabolism, biosynthesis of alkaloids derived from histidine and purine, and bile secretion pathways in untreated GD patients. In addition, pathways such as ABC transporters, folate biosynthesis, D-amino acid metabolism, anthranilate degradation, and purine metabolism remained significantly dysregulated after treatment. Correlation analysis revealed positive associations between docosatetraenoic acid and citric acid with free triiodothyronine and free thyroxine levels, between 4-hydroxyphenyllactic acid and free triiodothyronine, and between 13Z,16Z-docosadienoic acid and thyroid-stimulating hormone receptor antibody levels.
Conclusion: Significant alterations in plasma metabolomic profiles during the transition from hyperthyroidism to euthyroidism in GD patients were identified using untargeted metabolomics, highlighting persistent metabolic disruptions despite clinical recovery.
Keywords: Graves’ disease; biomarkers; metabolic pathway; untargeted metabolomics.
Conflict of interest statement
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the work reported.
Figures


References
LinkOut - more resources
Full Text Sources

