Inhibition of virally induced TFEB proteasomal degradation as a host-centric therapeutic approach for coronaviral infection
- PMID: 40465712
- PMCID: PMC12136031
- DOI: 10.1126/sciadv.adv4033
Inhibition of virally induced TFEB proteasomal degradation as a host-centric therapeutic approach for coronaviral infection
Abstract
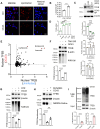
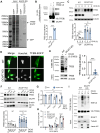
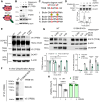
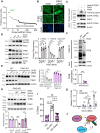
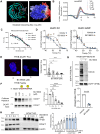
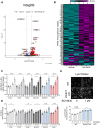
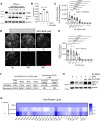
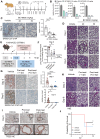
The endolysosomal pathway plays an evolutionarily conserved role in pathogen clearance, and viruses have evolved complex mechanisms to evade this host defense system. Here, we describe a previously unidentified aspect of coronaviral infection, whereby the master transcriptional activator of lysosomal homeostasis-TFEB-is targeted for proteasomal-mediated degradation upon viral infection. Through mass spectrometry analysis and an unbiased small interfering RNA screen, we identify that TFEB protein stability is coordinately regulated by the E3 ubiquitin ligase subunit DCAF7 and the PAK2 kinase. We derive a series of novel small molecules that interfere with the DCAF7-TFEB interaction. These agents inhibit virus-induced TFEB degradation and demonstrate broad antiviral activities including attenuating severe acute respiratory syndrome coronavirus 2 infection in two animal models. Together, these results delineate a virally triggered pathway that impairs lysosomal homeostasis in the host. Small molecule E3 ubiquitin ligase DCAF7 inhibitors that restore lysosomal function represent a novel class of host-directed, antiviral therapies useful for current and potentially future coronaviral variants.
Figures
References
-
- World Health Organization, WHO Coronavirus Disease (COVID-19) Dashboard (2024); https://data.who.int/dashboards/covid19/.
-
- Merad M., Blish C. A., Sallusto F., Iwasaki A., The immunology and immunopathology of COVID-19. Science 375, 1122–1127 (2022). - PubMed
-
- Wang X., Lu L., Jiang S., SARS-CoV-2 evolution from the BA.2.86 to JN.1 variants: Unexpected consequences. Trends Immunol. 45, 81–84 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

