Fecal metabolite profiling identifies critically ill patients with increased 30-day mortality
- PMID: 40465720
- PMCID: PMC12136028
- DOI: 10.1126/sciadv.adt1466
Fecal metabolite profiling identifies critically ill patients with increased 30-day mortality
Abstract
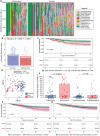
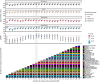
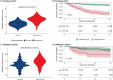
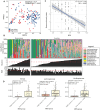
Critically ill patients admitted to the medical intensive care unit (MICU) have reduced intestinal microbiota diversity and altered microbiome-associated metabolite concentrations. Metabolites produced by the gut microbiota have been associated with survival of patients receiving complex medical treatments and thus might represent a treatable trait to improve clinical outcomes. We prospectively collected fecal specimens, defined microbiome compositions by shotgun metagenomic sequencing, and quantified microbiota-derived fecal metabolites by mass spectrometry from 196 critically ill patients admitted to the MICU for non-COVID-19 respiratory failure or shock to correlate microbiota features and metabolites with 30-day mortality. Microbiota compositions of the first fecal sample after MICU admission did not independently associate with 30-day mortality. We developed a metabolic dysbiosis score (MDS) that uses fecal concentrations of 13 microbiota-derived metabolites, which predicted 30-day mortality independent of known confounders. The MDS complements existing tools to identify patients at high risk of mortality by incorporating potentially modifiable, microbiome-related, independent contributors to host resilience.
Figures

References
-
- Maslove D. M., Tang B., Shankar-Hari M., Lawler P. R., Angus D. C., Baillie J. K., Baron R. M., Bauer M., Buchman T. G., Calfee C. S., Dos Santos C. C., Giamarellos-Bourboulis E. J., Gordon A. C., Kellum J. A., Knight J. C., Leligdowicz A., McAuley D. F., McLean A. S., Menon D. K., Meyer N. J., Moldawer L. L., Reddy K., Reilly J. P., Russell J. A., Sevransky J. E., Seymour C. W., Shapiro N. I., Singer M., Summers C., Sweeney T. E., Thompson B. T., van der Poll T., Venkatesh B., Walley K. R., Walsh T. S., Ware L. B., Wong H. R., Zador Z. E., Marshall J. C., Redefining critical illness. Nat. Med. 28, 1141–1148 (2022). - PubMed
-
- Agudelo-Ochoa G. M., Valdés-Duque B. E., Giraldo-Giraldo N. A., Jaillier-Ramírez A. M., Giraldo-Villa A., Acevedo-Castaño I., Yepes-Molina M. A., Barbosa-Barbosa J., Benítez-Paéz A., Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes 12, 1707610 (2020). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

