Epithelial GREMLIN1 disrupts intestinal epithelial-mesenchymal crosstalk to induce a wnt-dependent ectopic stem cell niche through stromal remodelling
- PMID: 40467544
- PMCID: PMC12137559
- DOI: 10.1038/s41467-025-60364-6
Epithelial GREMLIN1 disrupts intestinal epithelial-mesenchymal crosstalk to induce a wnt-dependent ectopic stem cell niche through stromal remodelling
Abstract
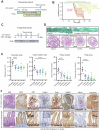
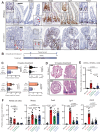
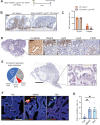
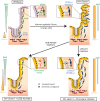
In homeostasis, counterbalanced morphogen signalling gradients along the vertical axis of the intestinal mucosa regulate the fate and function of epithelial and stromal cell compartments. Here, we use a disease-positioned mouse and human tissue to explore the consequences of pathological BMP signalling dysregulation on epithelial-mesenchymal interaction. Aberrant pan-epithelial expression of the secreted BMP antagonist Grem1 results in ectopic crypt formation, with lineage tracing demonstrating the presence of Lgr5(-) stem/progenitor cells. Isolated epithelial cell Grem1 expression has no effect on individual cell fate, indicating an intercompartmental impact of mucosal-wide BMP antagonism. Treatment with an anti-Grem1 antibody abrogates the polyposis phenotype, and triangulation of specific pathway inhibitors defines a pathological sequence of events, with Wnt-ligand-dependent ectopic stem cell niches forming through stromal remodelling following BMP disruption. These data support an emerging co-evolutionary model of intestinal cell compartmentalisation based on bidirectional regulation of epithelial-mesenchymal cell fate and function.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The named authors declare the following competing interests; S.J.L. has received grant income from UCB Pharma; G.D. was a UCB employee at the time the research was conducted; N.D. is an employee of UCB Pharma, UK and owns shares in UCB Pharma and Vertex Pharmaceuticals. All other authors declare no competing interests.
Figures



References
-
- Scoville, D., Sato, T., He, X. & Li, L. Current view: intestinal stem cells and signaling. Gastroenterology134, 849–864 (2008). - PubMed
-
- van den Brink, G. & Offerhaus, G. The morphogenetic code and colon cancer development. Cancer Cell11, 109–117 (2007). - PubMed
-
- McCarthy, N., Kraiczy, J. & Shivdasani, R. A. Cellular and molecular architecture of the intestinal stem cell niche. Nat. Cell Biol.22, 1033–1041 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

