Developing molecular surveillance of SARS-CoV-2 in the Czech Republic (2021-2022)
- PMID: 40467653
- PMCID: PMC12137580
- DOI: 10.1038/s41598-025-01074-3
Developing molecular surveillance of SARS-CoV-2 in the Czech Republic (2021-2022)
Abstract
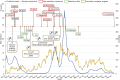
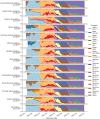
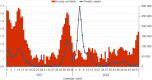
Molecular surveillance was widely used during the COVID-19 pandemic to detect rapidly emerging variants and monitor the transmission of SARS-CoV-2 within communities. In 2021, the Czech COVID-19 Genomics Consortium (COG-CZ) was set up to coordinate a new SARS-CoV-2 molecular surveillance network. In the Czech Republic, molecular surveillance employed whole genome sequencing (WGS) and variant discrimination polymerase chain reaction (VD-PCR) on samples collected through passive, active and sentinel surveillance. All WGS data was uploaded to GISAID and the PANGO lineages used by GISAID were compared to the main variants determined by VD-PCR. To assess the effectiveness and reliability of the gathered data in adapting pandemic responses, the capabilities and turnaround times of the molecular surveillance methods are evaluated. VD-PCR results were available within 48 h of sample collection for 81.5% of cases during the Delta/Omicron transition. WGS enabled the detection of low-frequency novel variants in infection clusters. WGS surveillance showed there was community spread of AY.20.1, a variant that gained novel mutations within the Czech Republic. Molecular surveillance informed the implementation of public health measures; temporal comparisons of restrictions and outcomes are described. Further areas for improvement have been identified for monitoring and managing future pandemics.
Keywords: Czech Republic; Molecular surveillance; SARS-CoV-2 variants; Variant discrimination PCR.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethical approval: Ethical approval was not necessary as data were collected as part of routine surveillance and only anonymised aggregated data were processed.
Figures




References
-
- Ministry of Health of the Czech Republic [Internet]. [cited 2023 Nov 10]. Available from: https://onemocneni-aktualne.mzcr.cz/covid-19.
-
- Institute of Health Information and Statistics of the Czech Republic. Podání žádosti O export, Analýzu Dat Z NZIS [Internet]. [cited 2023 Nov 10]. Available from: https://www.uzis.cz/index.php?pg=kontakt--zadosti-o-data-analyzy.
-
- Quick, J. NCoV-2019 Sequencing Protocol V3 (locost) [Internet]. protocols.io; 2020 [cited 2023 Nov 10]. Available from: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bp....
MeSH terms
Supplementary concepts
Grants and funding
- ECDC/HERA/2021/004 ECD.12218/European Centre for Disease Prevention and Control
- HERA2CZ/European Commission
- 101102733/European Commission
- CZ.02.1.01/0.0/0.0/16_019/0000785/European Commission
- ELIXIR CZ, LM2023055/Ministry of Education, Youth and Sports of the Czech Republic
- ELIXIR CZ, LM2023055/Ministry of Education, Youth and Sports of the Czech Republic
- ELIXIR CZ, LM2023055/Ministry of Education, Youth and Sports of the Czech Republic
- ELIXIR CZ, LM2023055/Ministry of Education, Youth and Sports of the Czech Republic
- ELIXIR CZ, LM2023055/Ministry of Education, Youth and Sports of the Czech Republic
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

