Individual variations in glycemic responses to carbohydrates and underlying metabolic physiology
- PMID: 40467897
- PMCID: PMC12283382
- DOI: 10.1038/s41591-025-03719-2
Individual variations in glycemic responses to carbohydrates and underlying metabolic physiology
Abstract
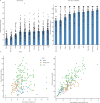
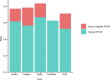
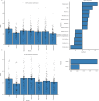
Elevated postprandial glycemic responses (PPGRs) are associated with type 2 diabetes and cardiovascular disease. PPGRs to the same foods have been shown to vary between individuals, but systematic characterization of the underlying physiologic and molecular basis is lacking. We measured PPGRs using continuous glucose monitoring in 55 well-phenotyped participants challenged with seven different standard carbohydrate meals administered in replicate. We also examined whether preloading a rice meal with fiber, protein or fat ('mitigators') altered PPGRs. We performed gold-standard metabolic tests and multi-omics profiling to examine the physiologic and molecular basis for interindividual PPGR differences. Overall, rice was the most glucose-elevating carbohydrate meal, but there was considerable interindividual variability. Individuals with the highest PPGR to potatoes (potato-spikers) were more insulin resistant and had lower beta cell function, whereas grape-spikers were more insulin sensitive. Rice-spikers were more likely to be Asian individuals, and bread-spikers had higher blood pressure. Mitigators were less effective in reducing PPGRs in insulin-resistant as compared to insulin-sensitive participants. Multi-omics signatures of PPGR and metabolic phenotypes were discovered, including insulin-resistance-associated triglycerides, hypertension-associated metabolites and PPGR-associated microbiome pathways. These results demonstrate interindividual variability in PPGRs to carbohydrate meals and mitigators and their association with metabolic and molecular profiles.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.P.S. is a cofounder, scientific advisor and shareholder of Filtricine, Iollo, January AI, Marble Therapeutics, Next Thought AI, Personalis, Protos Biologics, Qbio, RTHM, SensOmics. M.P.S. is a scientific advisor and equity holder of Abbratech, Applied Cognition, Enovone, M3 Helium, Onza. M.P.S. is a scientific advisor and stock option holder of Jupiter Therapeutics, Mitrix, Neuvivo, Sigil Biosciences, WndrHLTH, Yuvan Research. M.P.S. is a cofounder and stock option holder of Crosshair Therapeutics. M.P.S. is an investor in and scientific advisor of R42 and Swaza. M.P.S. is an investor in Repair Biotechnologies. M.P.S. is a cofounder, shareholder and director of Exposomics, Fodsel, InVu Health. M.P.S. is a cofounder and equity holder of Mirvie, NiMo Therapeutics, Orange Street Ventures. A.A.M. is currently an employee of Google. D.P. and T.M. are members of the scientific advisory board of January AI. The other authors declare no competing interests.
Figures












References
-
- Klil-Drori, A. J., Azoulay, L. & Pollak, M. N. Cancer, obesity, diabetes, and antidiabetic drugs: is the fog clearing? Nat. Rev. Clin. Oncol.14, 85–99 (2017). - PubMed
-
- Retnakaran, R. et al. Risk factors for renal dysfunction in type 2 diabetes: U.K. Prospective Diabetes Study 74. Diabetes55, 1832–1839 (2006). - PubMed
-
- Klein, R., Klein, B. E., Moss, S. E., Davis, M. D. & DeMets, D. L. The Wisconsin epidemiologic study of diabetic retinopathy. III. Prevalence and risk of diabetic retinopathy when age at diagnosis is 30 or more years. Arch. Ophthalmol.102, 527–532 (1984). - PubMed
MeSH terms
Substances
Grants and funding
- F32DK126287/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- P30DK116074/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01 DK110186-01/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- F32 DK126287/DK/NIDDK NIH HHS/United States
- R01 DK139472/DK/NIDDK NIH HHS/United States
- 2T15LM007033/U.S. Department of Health & Human Services | NIH | U.S. National Library of Medicine (NLM)
- 11-23-PDF-76/American Diabetes Association (ADA)
- T15 LM007033/LM/NLM NIH HHS/United States
- R01 DK114183/DK/NIDDK NIH HHS/United States
- R01 DK110186/DK/NIDDK NIH HHS/United States
- UM1 TR004921/TR/NCATS NIH HHS/United States
- P30 DK116074/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

