Loss of colonic fidelity enables multilineage plasticity and metastasis
- PMID: 40468074
- PMCID: PMC12350155
- DOI: 10.1038/s41586-025-09125-5
Loss of colonic fidelity enables multilineage plasticity and metastasis
Abstract
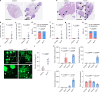
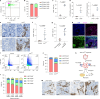
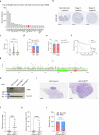
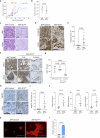
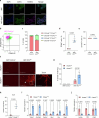
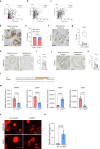
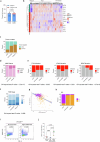
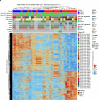
Cancer cell plasticity enables the acquisition of new phenotypic features and is implicated as a major driver of metastatic progression1,2. Metastasis occurs mostly in the absence of additional genetic alterations3-5, which suggests that epigenetic mechanisms are important6. However, they remain poorly defined. Here we identify the chromatin-remodelling enzyme ATRX as a key regulator of colonic lineage fidelity and metastasis in colorectal cancer. Atrx loss promotes tumour invasion and metastasis, concomitant with a loss of colonic epithelial identity and the emergence of highly plastic mesenchymal and squamous-like cell states. Combined analysis of chromatin accessibility and enhancer mapping identified impairment of activity of the colonic lineage-specifying transcription factor HNF4A as a key mediator of these observed phenotypes. We identify squamous-like cells in human patient samples and a squamous-like expression signature that correlates with aggressive disease and poor patient prognosis. Collectively, our study defines the epigenetic maintenance of colonic epithelial identity by ATRX and HNF4A as suppressors of lineage plasticity and metastasis in colorectal cancer.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

