CREM is a regulatory checkpoint of CAR and IL-15 signalling in NK cells
- PMID: 40468083
- PMCID: PMC12286855
- DOI: 10.1038/s41586-025-09087-8
CREM is a regulatory checkpoint of CAR and IL-15 signalling in NK cells
Abstract
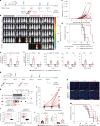
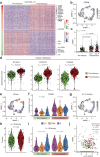
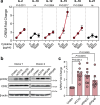
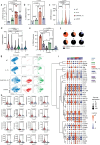
Chimeric antigen receptor (CAR) natural killer (NK) cell immunotherapy offers a promising approach against cancer1-3. However, the molecular mechanisms that regulate CAR-NK cell activity remain unclear. Here we identify the transcription factor cyclic AMP response element modulator (CREM) as a crucial regulator of NK cell function. Transcriptomic analysis revealed a significant induction of CREM in CAR-NK cells during the peak of effector function after adoptive transfer in a tumour mouse model, and this peak coincided with signatures of both activation and dysfunction. We demonstrate that both CAR activation and interleukin-15 signalling rapidly induce CREM upregulation in NK cells. Functionally, CREM deletion enhances CAR-NK cell effector function both in vitro and in vivo and increases resistance to tumour-induced immunosuppression after rechallenge. Mechanistically, we establish that induction of CREM is mediated by the PKA-CREB signalling pathway, which can be activated by immunoreceptor tyrosine-based activation motif signalling downstream of CAR activation or by interleukin-15. Finally, our findings reveal that CREM exerts its regulatory functions through epigenetic reprogramming of CAR-NK cells. Our results provide support for CREM as a therapeutic target to enhance the antitumour efficacy of CAR-NK cells.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.R., R.B., S.A., M.S., M. Daher, P.B., N.U., Y.L., P. Lin, D.M., E.J.S. and K.R. and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Takeda Pharmaceutical. S.A., R.B., D.M., E.J.S. and K.R. have an institutional financial conflict of interest with Affimed. A.M. is listed as an inventor on a patent that has been licensed by Johns Hopkins University to ThriveEarlier Detection. A.M. serves as a consultant for Tezcat Biotechnology. A.M. receives royalties from a patent that is licensed to Exact Sciences. K.R. participates on the Scientific Advisory Board for Avenge Bio, Virogin Biotech, Navan Technologies, Caribou Biosciences, Bit Bio, Replay, oNKo Innate, The Alliance for Cancer Gene Therapy ACGT, Innate Pharma and Shinobi Therapeutics. K.R. is the scientific founder of Syena. E.J.S. participates on the Scientific Advisory Board for Adaptimmune Limited, Axio Research, Celaid Therapeutics, FibroBiologics, Navan Technologies, New York Blood Center and Zelluna Immunotherapy. M. Daher participates on the Scientific Advisory Board of Cellsbin. The remaining authors declare that they have no competing interests.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

