PpMYC2 and PpJAM2/3 antagonistically regulate lignin synthesis to cope with the disease in peach fruit
- PMID: 40468145
- PMCID: PMC12392935
- DOI: 10.1111/pbi.70177
PpMYC2 and PpJAM2/3 antagonistically regulate lignin synthesis to cope with the disease in peach fruit
Abstract
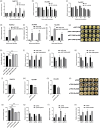
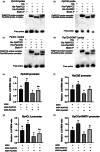
Transcription factors MYC2 and JAMs play a crucial role in regulating disease resistance in model plants. However, their regulatory mechanisms of disease resistance in non-model plants, particularly in postharvest fruits, remain largely unknown. In this study, we observed that PpMYC2 expression was up-regulated in peach fruit following infection by Monilinia fructicola, while the expressions of PpJAM2/3 were down-regulated. Furthermore, we found that PpMYC2 positively regulated the resistance against M. fructicola, whereas PpJAM2/3 negatively regulated it. Through a combined DNA affinity purification and RNA sequencing analysis for PpMYC2, we identified lignin synthesis genes (PpPAL1, PpC4H, Pp4CL1, PpCSE and PpCCoAOMT1) as candidate target genes. Subsequent assays, including dual-luciferase reporter assay, transient overexpression and silencing assays, electrophoretic mobility shift assay and yeast one-hybrid assay demonstrated that PpMYC2 activated the transcription of these five genes by binding to their promoters, promoting lignin accumulation. Conversely, PpJAM2 inhibited the transcription of PpC4H and PpCSE, while PpJAM3 inhibited Pp4CL1 and PpCCoAOMT1. Additionally, PpJAM2 or PpJAM3 interfered with PpMYC2's activation of their common target genes by competitively binding to the promoters. In conclusion, when peach fruit is infected with M. fructicola, up-regulation of PpMYC2 promotes lignin synthesis, while down-regulation of PpJAM2/3 reduces their inhibitory effects, ultimately resulting in lignin accumulation to combat the disease infection. Our study provides new insights into the molecular mechanisms of disease response in postharvest peach fruit.
Keywords: Monilinia fructicola; Prunus persica; competitive binding; lignin synthesis; transcription factor.
© 2025 The Author(s). Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Chen, J. , Wang, W. , Li, T. , Gao, X. , Zhang, X. and Zeng, K. (2023) CsAP2L transcription factor regulates resistance of citrus fruit to Penicillium digitatum through lignin biosynthesis and ROS metabolism pathways. Postharvest Biol. Technol. 205, 112499.
-
- Cheng, C. , Yan, C. , Qi, C. , Zhao, X. , Liu, L. , Guo, Y. , Leng, P. et al. (2023) Metabolome and transcriptome analysis of postharvest peach fruit in response to fungal pathogen Monilinia fructicola infection. Lwt‐Food Sci Technol. 173, 114301.
-
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

