The lactylation-immune regulatory axis: a potential therapeutic target for migraine prevention and treatment
- PMID: 40468199
- PMCID: PMC12139184
- DOI: 10.1186/s10194-025-02075-3
The lactylation-immune regulatory axis: a potential therapeutic target for migraine prevention and treatment
Abstract
Background: Migraine is a debilitating neurological disorder. Emerging evidence suggests that metabolic dysregulation and immune dysfunction contribute to migraine pathogenesis while the molecular mechanisms linking these processes remain unclear. Lactylation may serve as a crucial integrator of metabolic and immune signals in migraine.
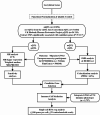
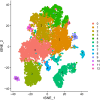
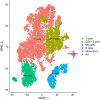
Methods: We performed a comprehensive multi-omics Mendelian randomization study integrating DNA methylation, gene expression, and protein abundance data with genome-wide association studies on migraine. Summary-data-based Mendelian randomization, Bayesian colocalization, and two-sample MR analyses were conducted to identify lactylation-related genes causally associated with migraine. We further explored immune cell mediation using genetic data from 731 immune phenotypes and validated findings through single-cell RNA sequencing of peripheral blood mononuclear cells from migraine patients and controls.
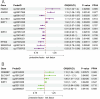
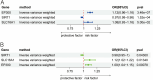
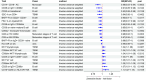
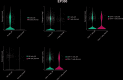
Results: EP300, SIRT1, and SLC16A1 were identified as key regulators of migraine susceptibility across methylation, expression, and protein levels. EP300 and SLC16A1 were associated with increased migraine risk, while SIRT1 conferred a protective effect. Mediation analyses revealed that genetic effects of these genes were partially transmitted through specific immune cell subsets, particularly B cells and natural killer T cells. Single-cell transcriptomic profiling further demonstrated EP300 upregulation in B cells and T cells of migraine patients. These findings support a novel “lactylation–immune mediation–migraine axis” linking metabolic and immune dysregulation to migraine pathogenesis.
Conclusion: This integrative multi-omics analysis uncovers lactylation-related genes as causal drivers of migraine through immunometabolic pathways. Targeting lactylation-regulated metabolic and immune mechanisms may offer novel precision therapeutic strategies for migraine, particularly in patients with inflammatory or metabolic endophenotypes.
Supplementary Information: The online version contains supplementary material available at 10.1186/s10194-025-02075-3.
Keywords: Immune mediation; Lactylation; Mendelian randomization; Migraine; Multi-omics; Single-cell RNA sequencing.
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
-
- Sutherland HG, Jenkins B, Griffiths LR (2024) Genetics of migraine: complexity, implications, and potential clinical applications. Lancet Neurol 23(4):429–446 - PubMed
-
- Khan J, Asoom LIA, Sunni AA, Rafique N, Latif R, Saif SA, Almandil NB, Almohazey D, AbdulAzeez S, Borgio JF (2021) Genetics, pathophysiology, diagnosis, treatment, management, and prevention of migraine. Biomed pharmacotherapy = Biomedecine Pharmacotherapie 139:111557 - PubMed
-
- Safiri S, Pourfathi H, Eagan A, Mansournia MA, Khodayari MT, Sullman MJM, Kaufman J, Collins G, Dai H, Bragazzi NL et al (2022) Global, regional, and National burden of migraine in 204 countries and territories, 1990 to 2019. Pain 163(2):e293–e309 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

