Integrated transcriptomic and metabolomic analyses reveal tissue-specific accumulation and expression patterns of monoterpene glycosides, gallaglycosides, and flavonoids in Paeonia Lactiflora Pall
- PMID: 40468204
- PMCID: PMC12139386
- DOI: 10.1186/s12864-025-11750-3
Integrated transcriptomic and metabolomic analyses reveal tissue-specific accumulation and expression patterns of monoterpene glycosides, gallaglycosides, and flavonoids in Paeonia Lactiflora Pall
Abstract
Background: Paeonia lactiflora Pall. (PL) is widely recognized for its ornamental, edible, and medicinal properties. Its principle bioactive constituents include monoterpene glycosides (MGs), gallaglycosides (GGs), and flavonoids. However, the metabolic and molecular basis underlying their biosynthesis in PL remain poorly understood. In this study, an integrated non-targeted metabolomics and transcriptomics approach was employed to investigate the metabolic profiles and gene expression patterns in four distinct PL tissues.
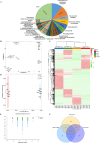
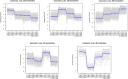
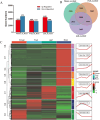
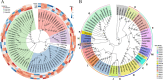
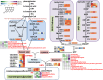
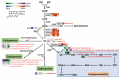
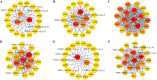
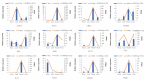
Results: Metabolomic and transcriptome profiling revealed tissue-specific patterns of metabolite accumulation and gene expression. KEGG enrichment analysis of differentially expressed metabolites (DEMs) showed that secondary metabolites biosynthesis and transport processes play vital roles in the tissue-specific accumulation of bioactive constituents. A total of 19 DEMs and 90 differentially expressed genes (DEGs) associated with MGs, 10 DEMs and 14 DEGs associated with GGs, and 205 DEMs and 67 DEGs associated with flavonoids were identified. Roots, the primary medicinal tissue, exhibited substantial accumulation of eight MGs, two GGs, and 18 flavonoids, as well as elevated expression levels of 16, two and nine structural genes, respectively. Nine CYP450 s and two UGTs associated with MGs, and 14 UGTs associated with flavonoids, were identified as new candidate genes through phylogenetic and expression analyses. CYP71E1, CYP71 AN24.1, CYP71 AU50.2, and UGT91 A1.1 for MGs biosynthesis, and UGT71 K1.4, UGT89B2, UGT73 C25, and UGT71 K1.2 for flavonoids biosynthesis were prioritized through correlation analysis. WGCNA revealed that turquoise, green, and blue modules were significantly correlated with MGs and flavonoids biosynthesis, identifying 24 hub genes for MGs and 18 for flavonoids. The overlap of phylogenetic, expression, correlation and WGCNA analyses identified CYP71 AN24.1 and UGT91 A1.1 as putative MGs biosynthetic genes, and UGT89B2 as a flavonoid-related gene. Protein structure prediction and similarity analysis further supported their functional conservation with known terpenoid-modifying enzymes and flavonoid-specific glycosyltransferases, respectively.
Conclusions: These findings identified CYP71 AN24.1, UGT91 A1.1, and UGT89B2 as novel genes involved in MGs and flavonoids biosynthesis. The study provides a valuable theoretical foundation for future metabolic engineering aimed at optimizing the biosynthetic pathways of these primary active constituents in PL.
Keywords: Paeonia Lactiflora; Biosynthesis; Flavonoids; Gallaglycoside; Metabolomic; Monoterpene glycosides; Transcriptome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






References
-
- Parker S, May B, Zhang C, Zhang AL, Lu C, Xue CC. A pharmacological review of bioactive constituents of Paeonia lactiflora Pall. and Paeonia veitchii Lynch. Phytother Res. 2016;30:1445–73. 10.1002/ptr.5653. - PubMed
-
- Ma X, Wen JX, Gao SJ, He X, Li PY, Yang YX, et al. Paeonia lactiflora Pall. regulates the NF-κB-NLRP3 inflammasome pathway to alleviate cholestasis in rats. J Pharma Pharmacol. 2018;70:1675–87. 10.1111/jphp.13008. - PubMed
-
- Lee HS, Kim Y. Paeonia lactiflora inhibits cell wall synthesis and triggers membrane depolarization in Candida albicans. J Microbiol Biotechnol. 2017;27:395–404. 10.4014/jmb.1611.11064. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

