Comparative study on the mechanism of yellow petal formation in Paphiopedilum armeniacum: an integrated transcriptomic and metabolomic analysis of three Paphiopedilum species
- PMID: 40468222
- PMCID: PMC12135254
- DOI: 10.1186/s12864-025-11648-0
Comparative study on the mechanism of yellow petal formation in Paphiopedilum armeniacum: an integrated transcriptomic and metabolomic analysis of three Paphiopedilum species
Abstract
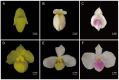
Backgrounds: Paphiopedilum orchids, particularly the Chinese endemic Paphiopedilum armeniacum, are prized for their commercial and ornamental value, with the latter serving as a vital breeding resource owing to its distinctive yellow petals. However, the molecular mechanisms underlying yellow petal formation remain unclear.
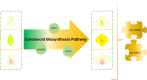
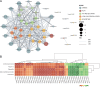
Results: This work employed an integrated transcriptomic and metabolomic comparative analysis of P. armeniacum and two lighter-colored Paphiopedilum species (The sepals and petals are white.) to identify carotenoid-related differentially expressed genes and metabolites before and after blooming in all three species. Metabolomic analysis revealed a marked increase in six differential metabolites, including zeaxanthin, precorrin 2, and β-D-gentiobiosyl crocetin, in P. armeniacum, highlighting their critical role in yellow petal formation. Transcriptomic comparison identified 40 DEGs (including D27, GDSL-like, CYP97B3, LUT1, and PSY) linked to yellow pigmentation, most of which were consistently upregulated in P. armeniacum before and after blooming Integrative metabolomic and transcriptomic analyses demonstrated significant correlations between these genes and metabolites, suggesting their role in regulating carotenoid synthesis and accumulation in yellow petal formation. Furthermore, qRT-PCR elucidates the expression levels of candidate genes, identifying RPL13AD as the optimal reference gene across these three orchid species.
Conclusions: These works elucidate the expression patterns and regulatory roles of carotenoid-related genes in metabolic pathways during P. armeniacum blooming, providing new insights into the molecular mechanisms of carotenoid-mediated plant coloration.
Keywords: Biochemical pathways; Carotenoids; Orchid; RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- Teoh ES. Orchids of Asia. 3rd ed. Singapore: Times Editions-Marshall Cavendish; 2005.
-
- Wang M, Li SZ, Chen LJ, Li J, Li LQ, Rao WH, et al. Conservation and reintroduction of the rare and endangered Orchid Paphiopedilum armeniacum. Ecosyst Health Sustain. 2021;7(1):1903817.
-
- Mou ZMM, Yang N, Li SY, Hu H. Nitrogen requirements for vegetative growth, blooming, seed production, and Ramet growth of Paphiopedilum Armeniacum (Orchid). HortScience. 2012;47:585–8.
-
- Chen XQ, Stephan W. Flora of China. Vol. 25, Orchidaceae. Beijing: Science; St. Louis: Botanical Garden Press;; 2009. pp. 36–7.
-
- Averyanov L, Cribb P, Loc PK, Hiep NT. Slipper orchids of Vietnam. Compass Press Limited; 2003.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

