GUIDE: a prospective cohort study for blood-based early detection of gastrointestinal cancers using targeted DNA methylation and fragmentomics sequencing
- PMID: 40468355
- PMCID: PMC12139136
- DOI: 10.1186/s12943-025-02367-x
GUIDE: a prospective cohort study for blood-based early detection of gastrointestinal cancers using targeted DNA methylation and fragmentomics sequencing
Abstract
Background: Gastrointestinal (GI) cancers are among the most prevalent and lethal malignancies worldwide. Early, non-invasive detection is essential for timely intervention and improved survival. To address this clinical need, we developed GutSeer, a blood-based assay combining DNA methylation and fragmentomics for multi-GI cancer detection.
Methods: Genome-wide methylome profiling identified 1,656 markers specific to five major GI cancers and their tissue origins. Based on these findings, we designed GutSeer, a targeted bisulfite sequencing panel, which was trained and validated using plasma samples from 1,057 cancer patients and 1,415 non-cancer controls. The locked model was blindly tested in an independent cohort of 846 participants, encompassing both inpatient and outpatient settings across five hospitals.
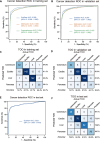
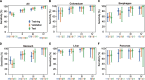
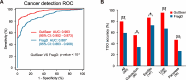
Results: In the validation cohort, GutSeer achieved an area under the curve (AUC) of 0.950 [95% Confidence Interval (CI): 0.937-0.962] for cancer detection, with 82.8% sensitivity (95% CI: 79.5-86.0) and 95.8% specificity (95% CI: 94.3-97.2). It detected 92.2% of colorectal, 75.5% of esophageal, 65.3% of gastric, 92.9% of liver, and 88.6% of pancreatic cancers. The independent test cohort included 198 early-stage cancers (stage I/II, 66.4%) and 63 advanced precancerous lesions. GutSeer maintained robust performance, with 81.5% sensitivity (95% CI: 77.1-85.9) for GI cancers and 94.4% specificity (95% CI: 92.4-96.5). It also demonstrated the ability to detect advanced precancerous lesions in the colorectum, esophagus, and stomach as a single, non-invasive blood test.
Conclusions: By integrating DNA methylation and fragmentomics into a compact panel, GutSeer outperformed genome-wide sequencing in both accuracy and clinical applicability. Its high sensitivity for early-stage GI cancers and practicality as a non-invasive assay highlights its potential to revolutionize early cancer detection and improve patient outcomes.
Trial registration: ClinicalTrials.gov identifier: NCT05431621.
Keywords: Cell-free DNA (cfDNA); Fragmentomics; Gastrointestinal cancer; Methylation; Multi-cancer early detection (MCED); Multi-dimensional features.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Ethical approval was obtained from the institutional review boards and the ethical committee of all participating centers: Zhongshan Hospital of Fudan University (No. B2020-291R), Changhai Hospital (No. CHEC2020-113), Qingpu Branch of Zhongshan Hospital affiliated to Fudan University (No. 2020-35), Xuhui Central Hospital (No. 2020-178), and Hubei Cancer Hospital (No. LCKY2021021). Informed consent was obtained from all participants before the study. This study was conducted in accordance with the Declaration of Helsinki and compliant with GCP guidelines. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Crosby D, Bhatia S, Brindle KM, Coussens LM, Dive C, Emberton M, Esener S, Fitzgerald RC, Gambhir SS, Kuhn P, et al. Early detection of cancer. Science. 2022;375:eaay9040. - PubMed
-
- Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, Kramer J, Siegel RL. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72:409–36. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- 82488101/National Natural Science Foundation of China
- 82341027/National Natural Science Foundation of China
- 82072715/National Natural Science Foundation of China
- SHDC2023CRD025/Shanghai hospital development center
- 201940075/Shanghai Municipal Health Commission
- 2022LJ005/Shanghai Municipal Health Commission
- 2021ZSCX28/Science Foundation of Zhongshan Hospital, Fudan University
- 2020ZSLC31/Science Foundation of Zhongshan Hospital, Fudan University
- 22S31901800/Shanghai Science and Technology Commission
- 21140900300/Shanghai Science and Technology Commission
- 2019YFC1315800/National Key Research and Development Program of China
- 2019YFC1315802/National Key Research and Development Program of China
- 2021YFC2501900/National Key Research and Development Program of China
- 2019YFC1315800/National Key Research and Development Program of China
- 81830102/State Key Program of the National Natural Science Foundation of China
- 82150004/Original Discovery Program of National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical

