LC-MS/MS proteomics identifies plasma proteins related to cognition over 9-year follow-up
- PMID: 40469059
- PMCID: PMC12138273
- DOI: 10.1002/alz.70276
LC-MS/MS proteomics identifies plasma proteins related to cognition over 9-year follow-up
Abstract
Introduction: This project identified plasma proteins predictive of cognitive decline across a robust neuropsychological protocol over a 9-year period.
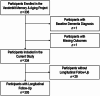
Methods: Vanderbilt Memory and Aging Project participants (n = 336, 73 ± 7 years, 59% male, 87% non-Hispanic White, 10% Black/African American) underwent blood draw for baseline plasma protein abundance using mass spectrometry analysis of tandem mass tag (TMT)-labeled peptides and serial neuropsychological assessment (follow-up = 6.1 ± 2.3 years). Linear mixed-effects regressions related protein levels to neuropsychological outcomes in fully adjusted models. False discovery rate correction was applied.
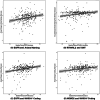
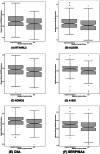
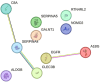
Results: Initial proteomics analyses yielded 3764 unique protein identifications across 23 16-plex TMT batches, and 686 proteins passed quality control. Proteins were identified predicting longitudinal decline in language (EGFR, RTN4RL2), information processing speed (EGFR, NOMO2, CLEC3B), executive function (A1BG), and visuospatial skills (RTN4RL2, GALNT1, SERPINA4, SERPINA5, C8A, ALDOB), but not episodic memory.
Discussion: Large-scale proteomics analyses identified 10 plasma proteins that predicted subsequent cognitive decline over a 9-year follow-up in multiple cognitive domains.
Highlights: There were 3764 unique protein identifications across 23 16-plex TMT batches. Rigorous quality control yielded 686 proteins used as predictors in analyses. Identified proteins related to all domains assessed, except for episodic memory. Many proteins identified were differentially expressed in MCI.
Keywords: Alzheimer's disease; LC‐MS/MS; cognition; mild cognitive impairment; plasma proteomics.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
T.J.H. serves on the Scientific Advisory Board for Vivid Genomics. T.J.H. is also Deputy Editor for
Figures






References
MeSH terms
Substances
Grants and funding
- FO2024-0048-TK-130/Hjärnfonden, Sweden
- ADSF-21-831381-C/AD Strategic Fund and the Alzheimer's Association
- 22HLT07/European Partnership on Metrology, co-financed from the European Union's Horizon Europe Research and Innovation Programme
- IIRG-08-88733/ALZ/Alzheimer's Association/United States
- ALFGBG-965240/Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement
- ALFGBG-1006418/Vetenskapsrådet
- AF-939721/Swedish Alzheimer Foundation
- #ALFGBG-71320/Swedish State Support for Clinical Research
- 22HLT07/HORIZON EUROPE European Research Council
- T32 AG058524/AG/NIA NIH HHS/United States
- R01 AG034962/AG/NIA NIH HHS/United States
- R01-AG064950/AG/NIA NIH HHS/United States
- F30-AG085905/AG/NIA NIH HHS/United States
- P20 AG068082/AG/NIA NIH HHS/United States
- 201809-2016862/Alzheimer's Drug Discovery Foundation
- R01-AG034962/AG/NIA NIH HHS/United States
- F31 AG079640/AG/NIA NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- AF-994551/Swedish Alzheimer Foundation
- 2022-00732/Swedish Research Council supported by grants from the Swedish Research Council
- 2023-00356/Swedish Research Council supported by grants from the Swedish Research Council
- K24-AG046373/AG/NIA NIH HHS/United States
- FO2024-0048-HK-24/Hjärnfonden, Sweden
- R01 AG056534/AG/NIA NIH HHS/United States
- ALZ2022-0006/Hjärnfonden, Sweden
- F31-AG079640/AG/NIA NIH HHS/United States
- ZEN-21-848495/Alzheimer's Association 2021 Zenith
- Vanderbilt University
- S10-OD023680/NIH Office of the Director
- UL1-TR002243/TR/NCATS NIH HHS/United States
- 2022-01018/Swedish Research Council supported by grants from the Swedish Research Council
- R01 AG064950/AG/NIA NIH HHS/United States
- P20-AG068082/AG/NIA NIH HHS/United States
- ALFGBG-1006418/Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement
- 101053962/European Union's Horizon Europe Research and Innovation Programme
- T32-GM007347/GM/NIGMS NIH HHS/United States
- PND2021-00694/European Union Joint Programme-Neurodegenerative Disease Research
- JPND2019-466-236/European Union Joint Program for Neurodegenerative Disorders
- R01-AG056534/AG/NIA NIH HHS/United States
- JPND2019-466-236/UK Dementia Research Institute
- UL1-TR000445/TR/NCATS NIH HHS/United States
- T32-AG058524/AG/NIA NIH HHS/United States
- UKDRI-1003/UK Dementia Research Institute at UCL
- 860197/Marie Skłodowska-Curie
- T32 GM007347/GM/NIGMS NIH HHS/United States
- 860197/HORIZON EUROPE Marie Sklodowska-Curie Actions
- ADSF-21-831376-C/AD Strategic Fund and the Alzheimer's Association
- S10 OD023680/OD/NIH HHS/United States
- FO2024-0048-HK-24/Hjärnfonden
- AF-994551/Alzheimerfonden
- ADSF-24-1284328-C/AD Strategic Fund and the Alzheimer's Association
- AF-968270/Swedish Alzheimer Foundation
- ADSF-21-831377-C/AD Strategic Fund and the Alzheimer's Association
- 2019-02397/Swedish Research Council supported by grants from the Swedish Research Council
- SG-23-1038904/Alzheimer's Association 2022-2025
- 201809-2016862/Alzheimer Drug Discovery Foundation (ADDF)
- UL1 TR002243/TR/NCATS NIH HHS/United States
- K24 AG046373/AG/NIA NIH HHS/United States
- AF-930351/Swedish Alzheimer Foundation
- FO2022-0270/Erling-Persson Family Foundation, Familjen Rönströms Stiftelse, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden
- 2017-00915/Swedish Research Council supported by grants from the Swedish Research Council
- F30 AG085905/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

