MAPK8 and HDAC6: potential biomarkers related to autophagy in diabetic retinopathy based on bioinformatics analysis
- PMID: 40469433
- PMCID: PMC12133481
- DOI: 10.3389/fendo.2025.1487007
MAPK8 and HDAC6: potential biomarkers related to autophagy in diabetic retinopathy based on bioinformatics analysis
Abstract
Introduction: One of the most common vascular diseases of the retina is diabetic retinopathy (DR), a microvascular condition caused by diabetes. The autophagy system transports and degrades cytoplasmic substances to lysosomes as part of the intracellular degradation process. Autophagy appears to be an important regulator in the development and progression of DR, but its mechanism and potential role are unclear. The purpose of this study is to identify autophagy-related genes in DR and find potential biomarkers associated with DR through bioinformatics analysis.
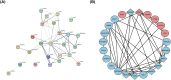
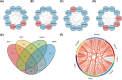
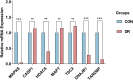
Method: We retrieved the dataset GSE102485 from the Gene Expression Omnibus (GEO) database and compiled a list of 344 autophagy-related genes. Using the R software, bioinformatics analysis was used to identify the differentially expressed autophagy-related genes (ARGs). Then, we identified the autophagy-related hub genes (ARHGs) through a series of analyses including Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, correlation analysis, and protein-protein interaction (PPI) network. In addition, the miRNA-gene-TF interaction network was generated using the NetworkAnalyst platform. Potential therapeutic drugs were predicted utilizing the Drug-Gene Interaction Database (DGIdb). Ultimately, DR was simulated through the high glucose incubation of the retinal pigment epithelium cell line (ARPE-19), and employing quantitative real-time polymerase chain reaction (qRT-PCR) to verify ARHG expression. The effectiveness of ARHGs in diagnosing DR was assessed by measuring the area under the receiver operating characteristic (ROC) curve.
Results: Differential expression analysis identified 26 ARGs, of which 6 were upregulated and 20 were downregulated. Through GO and KEGG enrichment analysis, it was found that ARGs showed significant enrichment in autophagy-related pathways. Using PPI network analysis, 7 ARHGs were identified. The expression of MAPK8, HDAC6, DNAJB1 and TARDBP, in a model of DR were confirmed by qRT-PCR. The ROC curve results showed that MAPK8, HDAC6, DNAJB1 and TSC2 had high predictive accuracy and could be used as biomarkers for DR.
Conclusion: Through bioinformatics analysis, we identified 26 genes that may be associated with autophagy in DR. We suggest that the hub genes MAPK8 and HDAC6 as biomarkers may be involved in autophagy in DR.
Keywords: HDAC6; MAPK8; autophagy; biomarker; diabetic retinopathy.
Copyright © 2025 Sun and Zuo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

