This is a preprint.
Semaphorin-7A promotes macrophage-mediated mammary epithelial and ductal carcinoma in situ invasion
- PMID: 40470186
- PMCID: PMC12136188
- DOI: 10.21203/rs.3.rs-6448305/v1
Semaphorin-7A promotes macrophage-mediated mammary epithelial and ductal carcinoma in situ invasion
Update in
-
Semaphorin-7A promotes macrophage-mediated mammary epithelial and ductal carcinoma in situ invasion.Breast Cancer Res. 2025 Dec 26;28(1):25. doi: 10.1186/s13058-025-02180-w. Breast Cancer Res. 2025. PMID: 41454324 Free PMC article.
Abstract
Background: Ductal carcinoma in situ (DCIS) accounts for 20-30% of all breast cancer diagnoses. Considered stage 0, DCIS is contained in the ducts by the myoepithelium that surround the luminal cells in the mammary gland. DCIS can progress to invasive ductal carcinoma (IDC) if the tumor cells break through the myoepithelium and invade the surrounding breast tissue. While 30-50% of DCIS tumors will progress to IDC, a majority will remain in a DCIS-like state. The mechanisms that drive this progression are not completely understood. There is currently no clinically recognized biomarker for predicting risk of DCIS progression. Therefore, all DCIS tumors are treated with standard of care, resulting in overtreatment. We have previously identified independent roles for semaphorin-7A (SEMA7A) and collagen in promoting DCIS progression to IDC.
Methods: To investigate the relationship between SEMA7A and collagen remodeling in the mammary gland, we utilized patient tissues and mouse models of normal development and DCIS progression as well as a novel SEMA7A-blocking antibody.
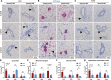
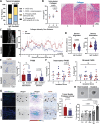
Results: We show that SEMA7A increases in patient samples of DCIS compared to matched normal tissues and in IDC compared to matched DCIS and normal tissues. This increase was correlated with the presence of CD68 + macrophages. Using puberty in the mammary gland as a model for normal epithelial invasion facilitated by macrophages, we show SEMA7A knockout mice exhibit delayed ductal elongation as well as decreased macrophages. Additionally, our SEMA7A-blocking antibody in a mouse model of DCIS decreased invasive tumor phenotypes and decreased organized collagen around the tumor. The invasive tumors had increased collagen and macrophage influx in the tumor. Finally, we show that SEMA7A activates an AKT/GSK3β/β-catenin signaling pathway within macrophages to promote expression of pro-inflammatory cytokines and the matrix remodeling enzyme MMP9 to facilitate invasion.
Conclusions: Our results demonstrate that SEMA7A regulates normal and transformed epithelial cell invasion through regulation of pro-invasive matrix remodeling via macrophages. Our studies also suggest that SEMA7A expression, macrophage phenotype, and collagen structure may be a predictor of risk for DCIS invasion. Thus, blocking SEMA7A may be a novel therapeutic strategy for high-risk DCIS patients to slow or prevent progression of disease.
Keywords: DCIS; ECM; SEMA7A; invasion; macrophage; matrix-remodeling.
Conflict of interest statement
Declarations Competing interests: TRL holds patent for SmAbH1. Other authors declare that they have no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

