Understanding the transition from embryogenesis to seed filling in Phaseolus vulgaris L. non-endospermic seeds
- PMID: 40470360
- PMCID: PMC12133513
- DOI: 10.3389/fpls.2025.1597915
Understanding the transition from embryogenesis to seed filling in Phaseolus vulgaris L. non-endospermic seeds
Abstract
Introduction: Common bean (Phaseolus vulgaris L.) is one of the most consumed grain legumes. These legumes are a major source of proteins and other important nutrients, especially in developing countries. Studying seed development in common bean is crucial for improving yield, nutrition, stress tolerance and disease resistance while promoting sustainable agriculture and food security, with its sequenced genome and available molecular tools making it an excellent research model. Despite advances in studying P. vulgaris seed development, the precise timing and molecular regulation of the transition from embryogenesis to seed filling remain poorly understood. Although P. vulgaris seeds at 10 days after anthesis (DAA) were previously characterized as being in the late embryogenesis stage, our previous studies suggested that this transition might occur earlier than 10 DAA, prompting us to investigate earlier developmental stages.
Methods: To accomplish this goal, we conducted a comprehensive analysis at 6, 10, 14, 18 and 20 DAA, integrating morphological, histological, and transcriptomic approaches.
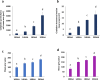
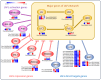
Results and discussion: Morphological and histochemical data revealed that by 10 DAA, cotyledons are fully formed, but storage compound accumulation is only noticed at 14 DAA, indicating that the transition from embryogenesis to seed filling occurs between 10 and 14 DAA. Transcriptomic analysis further supported this finding, showing upregulation of genes associated with seed storage proteins, starch metabolism, and hormonal regulation at 14 and 18 DAA. This study redefines the developmental timeline of P. vulgaris seed filling initiation, bridging a critical knowledge gap in legume seed biology. Given the limited availability of histological studies on early P. vulgaris seed development, our findings provide essential insights into the structural and molecular events driving this transition. By refining the timing and regulatory mechanisms of early seed development, this study lays the groundwork for future research aimed at enhancing seed quality and resilience in legumes.
Keywords: Phaseolus vulgaris L.; early seed filling; seed histology; storage compounds; transcriptome.
Copyright © 2025 Lopes, Fevereiro and Araújo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures






References
-
- Aubry C., Morère-Le Paven M. C., Chateigner-Boutin A. L., Teulat-Merah B., Ricoult C., Peltier D., et al. (2003). A gene encoding a germin-like protein, identified by a cDNA-AFLP approach, is specifically expressed during germination of Phaseolus vulgaris . Planta 217, 466–475. doi: 10.1007/S00425-003-1004-9/FIGURES/8 - DOI - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Society: Ser. B (Methodological) 57, 289–300. doi: 10.1111/J.2517-6161.1995.TB02031.X - DOI
-
- Bewley J. D., Bradford K. J., Hilhorst H. W. M., Nonogaki H. (2013). Seeds: Physiology of Development, Germination and Dormancy, 3rd edition. New York, NY: Springer, 1–392. doi: 10.1007/978-1-4614-4693-4 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

