This is a preprint.
Coexistence vs collapse in transposon populations
- PMID: 40470469
- PMCID: PMC12136480
Coexistence vs collapse in transposon populations
Update in
-
Coexistence versus collapse in transposon populations.Phys Rev E. 2025 May;111(5-1):054414. doi: 10.1103/PhysRevE.111.054414. Phys Rev E. 2025. PMID: 40534062
Abstract
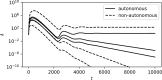
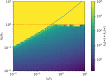
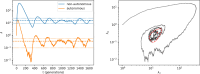
Transposons are small, self-replicating DNA sequences found in every branch of life. Often, one transposon will parasitize another, forming a tiny intracellular ecosystem. In some species these ecosystems thrive, while in others they go extinct, yet little is known about when or why this occurs. Here, we present a stochastic model for these ecosystems and discover a transition from stable coexistence to population collapse when the propensity for a transposon to replicate comes to exceed that of its parasites. Our model also predicts that replication rates should be low in equilibrium, which appears to be true of many transposons in nature.
Figures
References
-
- Siguier P., Gourbeyre E., and Chandler M., Bacterial insertion sequences: their genomic impact and diversity, FEMS Microbiology Reviews 38, 865 (2014), https://academic.oup.com/femsre/article-pdf/38/5/865/18142951/38-5-865.pdf. - PMC - PubMed
-
- Thomas J. and Pritham E. J., Helitrons, the eukaryotic rolling-circle transposable elements, Microbiology Spectrum 3, 10.1128/microbiolspec.mdna3 (2015), https://journals.asm.org/doi/pdf/10.1128/microbiolspec.mdna3-0049-2014. - DOI - PubMed
-
- Lander E., Linton L., Birren B., Nusbaum C., Zody M., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann Y., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R., Wilson R., Hillier L., McPherson J., Marra M., Mardis E., Fulton L., Chinwalla A., Pepin K., Gish W., Chissoe S., Wendl M., Delehaunty K., Miner T., Delehaunty A., Kramer J., Cook L., Fulton R., Johnson D., Minx P., Clifton S., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R., Muzny D., Scherer S., Bouck J., Sodergren E., Worley K., Rives C., Gorrell J., Metzker M., Naylor S., Kucherlapati R., Nelson D., Weinstock G., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R., Federspiel N., Abola A., Proctor M., Myers R., Schmutz J., Dickson M., Grimwood J., Cox D., Olson M., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G., Athanasiou M., Schultz R., Roe B., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J., Bateman A., Batzoglou S., Birney E., Bork P., Brown D., Burge C., Cerutti L., Chen H., Church D., Clamp M., Copley R., Doerks T., Eddy S., Eichler E., Furey T., Galagan J., Gilbert J., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L., Jones T., Kasif S., Kaspryzk A., Kennedy S., Kent W., Kitts P., Koonin E., Korf I., Kulp D., Lancet D., Lowe T., McLysaght A., Mikkelsen T., Moran J., Mulder N., Pollara V., Ponting C., Schuler G., Schultz J., Slater G., Smit A., Stupka E., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y., Wolfe K., Yang S., Yeh R., Collins F., Guyer M., Peterson J., Felsenfeld A., Wetterstrand K., Patrinos A., Morgan M., de P., Jong J. Catanese, Osoegawa K., Shizuya H., Choi S, Chen Y., and Szustakowki J., Initial sequencing and analysis of the human genome, Nature 409(6822), 860 (2001) - PubMed
-
- Kazazian J., HH C. Wong, Youssoufian H., Scott A., Phillips D., and Antonarakis S., Haemophilia a resulting from de novo insertion of l1 sequences represents a novel mechanism for mutation in man, Nature 332(6160), 164 (1988) - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
