Heat shock protein 90 chaperone activity is required for hepatitis A virus replication
- PMID: 40470959
- PMCID: PMC12282101
- DOI: 10.1128/jvi.00502-25
Heat shock protein 90 chaperone activity is required for hepatitis A virus replication
Abstract
HSP90 heat shock chaperones are essential for maintaining cellular proteostasis, as well as the ATP-dependent folding and functional maturation of many viral proteins. As a result, inhibitors of HSP90 have broad antiviral activity, disrupting replication of many viruses at concentrations below those causing cytotoxicity. Among the Picornaviridae, HSP90 inhibitors block replication of multiple Enterovirus, Aphthovirus, and Cardiovirus species, in some cases, by preventing post-translational processing and assembly of P1 capsid proteins. Hepatitis A virus (HAV), classified within the genus Hepatovirus, has been suggested to be an exception among picornaviruses and to replicate independently of HSP90, possibly because its slow translational kinetics could facilitate co-translational folding and assembly of its capsid proteins. However, we show here that HAV replication is highly dependent upon HSP90, both in human hepatocyte-derived cell lines, in which the 50% inhibitory concentration of geldanamycin was 8.7-11.8 nM, and in vivo in Ifnar1-/- mice. Label-free proteomics experiments suggested that HSP90 interacts with capsid proteins or their precursors and may thus facilitate the folding and assembly of capsid proteins, as it does for enteroviruses and aphthoviruses. By contrast, there was no evidence for HSP90 interacting with any nonstructural protein, and HSP90 inhibitors did not impair 3Cpro proteolytic activity. Despite this, and in contrast to previous studies of enteroviruses and aphthoviruses, geldanamycin potently inhibited replication of a subgenomic HAV replicon. We conclude that HAV is no exception from the HSP90-dependent nature of other picornaviruses and indeed is more dependent on HSP90 than other picornaviruses for amplification of its genome.IMPORTANCEHepatitis A virus (HAV), a common cause of acute infectious hepatitis, has been reported to differ from other picornaviruses in not requiring heat shock protein HSP90 for efficient replication. However, we show here that productive HAV infection is highly dependent on HSP90 and that HAV replication is potently blocked both in cell culture and in vivo in the murine liver by chemical inhibitors of HSP90. Such inhibitors also disrupt the replication of a subgenomic HAV RNA replicon, indicating that HSP90 is required for the assembly of functional replication organelles. This highlights a key difference from other picornaviruses for which HSP90 is required primarily, if not exclusively, for the maturation of the P1 capsid proteins.
Keywords: ACC1; HD-PTP; HSP70; HSP90 inhibitor; antiviral; chaperone; hepatovirus; mouse model; picornavirus; quasi-enveloped virus; replicon.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

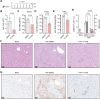



References
MeSH terms
Substances
Grants and funding
- K08-AI156125/National Institute of Allergy and Infectious Diseases
- K08 AI156125/AI/NIAID NIH HHS/United States
- R01 AI103083/AI/NIAID NIH HHS/United States
- R21 AG071229/AG/NIA NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- JP21fk0210053/Japan Agency for Medical Research and Development
- KAKENHI 24K10250/Japan Society for the Promotion of Science
- KAKENHI 24K09320/Japan Society for the Promotion of Science
- R41 DK133051/DK/NIDDK NIH HHS/United States
- R21-AG071229,R41-AG085859/AG/NIA NIH HHS/United States
- R01 AI150095/AI/NIAID NIH HHS/United States
- R41-DK133051/DK/NIDDK NIH HHS/United States
- R01-GM133107/GM/NIGMS NIH HHS/United States
- R01-AI103083,R01-AI150095/National Institute of Allergy and Infectious Diseases
- JP24fk0108627/Japan Agency for Medical Research and Development
- R41 AG085859/AG/NIA NIH HHS/United States
- R01 GM133107/GM/NIGMS NIH HHS/United States
- R21 AI163606/AI/NIAID NIH HHS/United States
- JP24fk0210109/Japan Agency for Medical Research and Development
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

